ISG15 Is a Novel Regulator of Lipid Metabolism during Vaccinia Virus Infection
- PMID: 36453897
- PMCID: PMC9769738
- DOI: 10.1128/spectrum.03893-22
ISG15 Is a Novel Regulator of Lipid Metabolism during Vaccinia Virus Infection
Abstract
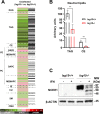
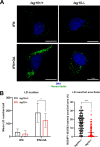
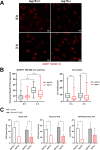
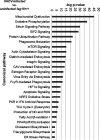
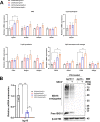
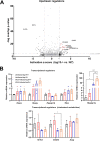
Interferon-stimulated gene 15 (ISG15) is a 15-kDa ubiquitin-like modifier that binds to _target proteins in a process termed ISGylation. ISG15, first described as an antiviral molecule against many viruses, participates in numerous cellular processes, from immune modulation to the regulation of genome stability. Interestingly, the role of ISG15 as a regulator of cell metabolism has recently gained strength. We previously described ISG15 as a regulator of mitochondrial functions in bone marrow-derived macrophages (BMDMs) in the context of Vaccinia virus (VACV) infection. Here, we demonstrate that ISG15 regulates lipid metabolism in BMDMs and that ISG15 is necessary to modulate the impact of VACV infection on lipid metabolism. We show that Isg15-/- BMDMs demonstrate alterations in the levels of several key proteins of lipid metabolism that result in differences in the lipid profile compared with Isg15+/+ (wild-type [WT]) BMDMs. Specifically, Isg15-/- BMDMs present reduced levels of neutral lipids, reflected by decreased lipid droplet number. These alterations are linked to increased levels of lipases and are independent of enhanced fatty acid oxidation (FAO). Moreover, we demonstrate that VACV causes a dysregulation in the proteomes of BMDMs and alterations in the lipid content of these cells, which appear exacerbated in Isg15-/- BMDMs. Such metabolic changes are likely caused by increased expression of the metabolic regulators peroxisome proliferator-activated receptor-γ (PPARγ) and PPARγ coactivator-1α (PGC-1α). In summary, our results highlight that ISG15 controls BMDM lipid metabolism during viral infections, suggesting that ISG15 is an important host factor to restrain VACV impact on cell metabolism. IMPORTANCE The functions of ISG15 are continuously expanding, and growing evidence supports its role as a relevant modulator of cell metabolism. In this work, we highlight how the absence of ISG15 impacts macrophage lipid metabolism in the context of viral infections and how poxviruses modulate metabolism to ensure successful replication. Our results open the door to new advances in the comprehension of macrophage immunometabolism and the interaction between VACV and the host.
Keywords: host-pathogen interactions; innate immunity; interferons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
ISG15 Is Required for the Dissemination of Vaccinia Virus Extracellular Virions.Microbiol Spectr. 2023 Jun 15;11(3):e0450822. doi: 10.1128/spectrum.04508-22. Epub 2023 Apr 10. Microbiol Spectr. 2023. PMID: 37036376 Free PMC article.
-
ISG15 regulates peritoneal macrophages functionality against viral infection.PLoS Pathog. 2013;9(10):e1003632. doi: 10.1371/journal.ppat.1003632. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24137104 Free PMC article.
-
ISG15 governs mitochondrial function in macrophages following vaccinia virus infection.PLoS Pathog. 2017 Oct 27;13(10):e1006651. doi: 10.1371/journal.ppat.1006651. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 29077752 Free PMC article.
-
Interferon-stimulated gene 15 and the protein ISGylation system.J Interferon Cytokine Res. 2011 Jan;31(1):119-30. doi: 10.1089/jir.2010.0110. Epub 2010 Dec 29. J Interferon Cytokine Res. 2011. PMID: 21190487 Free PMC article. Review.
-
The ISG15/USP18 ubiquitin-like pathway (ISGylation system) in hepatitis C virus infection and resistance to interferon therapy.Int J Biochem Cell Biol. 2011 Oct;43(10):1427-31. doi: 10.1016/j.biocel.2011.06.006. Epub 2011 Jun 16. Int J Biochem Cell Biol. 2011. PMID: 21704181 Review.
Cited by
-
Unveiling the Multifaceted Roles of ISG15: From Immunomodulation to Therapeutic Frontiers.Vaccines (Basel). 2024 Feb 1;12(2):153. doi: 10.3390/vaccines12020153. Vaccines (Basel). 2024. PMID: 38400136 Free PMC article. Review.
-
ISG15 Is Required for the Dissemination of Vaccinia Virus Extracellular Virions.Microbiol Spectr. 2023 Jun 15;11(3):e0450822. doi: 10.1128/spectrum.04508-22. Epub 2023 Apr 10. Microbiol Spectr. 2023. PMID: 37036376 Free PMC article.
-
Glycerol monolaurate inhibits Francisella novicida growth and is produced intracellularly in an ISG15-dependent manner.MicroPubl Biol. 2023 Oct 27;2023:10.17912/micropub.biology.000905. doi: 10.17912/micropub.biology.000905. eCollection 2023. MicroPubl Biol. 2023. PMID: 37954520 Free PMC article.
-
Stimulation of neutral lipid synthesis via viral growth factor signaling and ATP citrate lyase during vaccinia virus infection.J Virol. 2024 Nov 19;98(11):e0110324. doi: 10.1128/jvi.01103-24. Epub 2024 Oct 30. J Virol. 2024. PMID: 39475274 Free PMC article.
-
Lipid droplets in pathogen infection and host immunity.Acta Pharmacol Sin. 2024 Mar;45(3):449-464. doi: 10.1038/s41401-023-01189-1. Epub 2023 Nov 22. Acta Pharmacol Sin. 2024. PMID: 37993536 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

