Neutrophil profiles of pediatric COVID-19 and multisystem inflammatory syndrome in children
- PMID: 36476388
- PMCID: PMC9676175
- DOI: 10.1016/j.xcrm.2022.100848
Neutrophil profiles of pediatric COVID-19 and multisystem inflammatory syndrome in children
Abstract
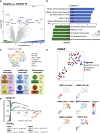
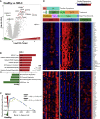
Multisystem inflammatory syndrome in children (MIS-C) is a delayed-onset, COVID-19-related hyperinflammatory illness characterized by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigenemia, cytokine storm, and immune dysregulation. In severe COVID-19, neutrophil activation is central to hyperinflammatory complications, yet the role of neutrophils in MIS-C is undefined. Here, we collect blood from 152 children: 31 cases of MIS-C, 43 cases of acute pediatric COVID-19, and 78 pediatric controls. We find that MIS-C neutrophils display a granulocytic myeloid-derived suppressor cell (G-MDSC) signature with highly altered metabolism that is distinct from the neutrophil interferon-stimulated gene (ISG) response we observe in pediatric COVID-19. Moreover, we observe extensive spontaneous neutrophil extracellular trap (NET) formation in MIS-C, and we identify neutrophil activation and degranulation signatures. Mechanistically, we determine that SARS-CoV-2 immune complexes are sufficient to trigger NETosis. Our findings suggest that hyperinflammatory presentation during MIS-C could be mechanistically linked to persistent SARS-CoV-2 antigenemia, driven by uncontrolled neutrophil activation and NET release in the vasculature.
Keywords: COVID-19; RNA sequencing; multisystem inflammatory syndrome in children; neutrophil; neutrophil extracellular traps; pediatrics.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.S.-F. receives funding from Bristol-Myers Squibb. G.A. is a founder of Seromyx Systems, Inc. A.F. is co-founder of and stockholder in Alba Therapeutics.
Figures

Update of
-
Neutrophil Profiles of Pediatric COVID-19 and Multisystem Inflammatory Syndrome in Children.bioRxiv [Preprint]. 2021 Dec 20:2021.12.18.473308. doi: 10.1101/2021.12.18.473308. bioRxiv. 2021. Update in: Cell Rep Med. 2022 Dec 20;3(12):100848. doi: 10.1016/j.xcrm.2022.100848 PMID: 34981052 Free PMC article. Updated. Preprint.
Similar articles
-
Neutrophil Profiles of Pediatric COVID-19 and Multisystem Inflammatory Syndrome in Children.bioRxiv [Preprint]. 2021 Dec 20:2021.12.18.473308. doi: 10.1101/2021.12.18.473308. bioRxiv. 2021. Update in: Cell Rep Med. 2022 Dec 20;3(12):100848. doi: 10.1016/j.xcrm.2022.100848 PMID: 34981052 Free PMC article. Updated. Preprint.
-
Long-term humoral signatures following acute pediatric COVID-19 and Multisystem Inflammatory Syndrome in Children.Pediatr Res. 2023 Oct;94(4):1327-1334. doi: 10.1038/s41390-023-02627-w. Epub 2023 May 12. Pediatr Res. 2023. PMID: 37173406 Free PMC article.
-
Measurement of Severe Acute Respiratory Syndrome Coronavirus 2 Antigens in Plasma of Pediatric Patients With Acute Coronavirus Disease 2019 or Multisystem Inflammatory Syndrome in Children Using an Ultrasensitive and Quantitative Immunoassay.Clin Infect Dis. 2022 Oct 12;75(8):1351-1358. doi: 10.1093/cid/ciac160. Clin Infect Dis. 2022. PMID: 35213684 Free PMC article.
-
Multisystem Inflammatory Syndrome in Children (MIS-C).Curr Allergy Asthma Rep. 2022 May;22(5):53-60. doi: 10.1007/s11882-022-01031-4. Epub 2022 Mar 22. Curr Allergy Asthma Rep. 2022. PMID: 35314921 Free PMC article. Review.
-
Neutrophils in COVID-19: Not Innocent Bystanders.Front Immunol. 2022 Jun 1;13:864387. doi: 10.3389/fimmu.2022.864387. eCollection 2022. Front Immunol. 2022. PMID: 35720378 Free PMC article. Review.
Cited by
-
COVID-19 mRNA vaccines induce robust levels of IgG but limited amounts of IgA within the oronasopharynx of young children.medRxiv [Preprint]. 2024 Apr 16:2024.04.15.24305767. doi: 10.1101/2024.04.15.24305767. medRxiv. 2024. Update in: J Infect Dis. 2024 Dec 16;230(6):1390-1399. doi: 10.1093/infdis/jiae450 PMID: 38699375 Free PMC article. Updated. Preprint.
-
Multisystem inflammatory syndrome in children characterized by enhanced antigen-specific T-cell expression of cytokines and its reversal following recovery.Front Pediatr. 2023 Dec 5;11:1235342. doi: 10.3389/fped.2023.1235342. eCollection 2023. Front Pediatr. 2023. PMID: 38116577 Free PMC article.
-
Plasma cell-free RNA signatures of inflammatory syndromes in children.Proc Natl Acad Sci U S A. 2024 Sep 10;121(37):e2403897121. doi: 10.1073/pnas.2403897121. Epub 2024 Sep 6. Proc Natl Acad Sci U S A. 2024. PMID: 39240972 Free PMC article.
-
Cardiac Involvement in Patients With Multisystem Inflammatory Syndrome in Adults.J Am Heart Assoc. 2024 Feb 20;13(4):e032143. doi: 10.1161/JAHA.123.032143. Epub 2024 Feb 13. J Am Heart Assoc. 2024. PMID: 38348793 Free PMC article. Review.
-
Circulating Spike Protein Detected in Post-COVID-19 mRNA Vaccine Myocarditis.Circulation. 2023 Mar 14;147(11):867-876. doi: 10.1161/CIRCULATIONAHA.122.061025. Epub 2023 Jan 4. Circulation. 2023. PMID: 36597886 Free PMC article.
References
-
- LaSalle T.J., Gonye A.L.K., Freeman S.S., Kaplonek P., Gushterova I., Kays K.R., Manakongtreecheep K., Tantivit J., Rojas-Lopez M., Russo B.C., et al. Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100779. - DOI - PMC - PubMed
-
- Aschenbrenner A.C., Mouktaroudi M., Krämer B., Oestreich M., Antonakos N., Nuesch-Germano M., Gkizeli K., Bonaguro L., Reusch N., Baßler K., et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021;13:7. doi: 10.1186/s13073-020-00823-5. - DOI - PMC - PubMed
-
- Meizlish M.L., Pine A.B., Bishai J.D., Goshua G., Nadelmann E.R., Simonov M., Chang C.H., Zhang H., Shallow M., Bahel P., et al. A neutrophil activation signature predicts critical illness and mortality in COVID-19. Blood Adv. 2021;5:1164–1177. doi: 10.1182/bloodadvances.2020003568. - DOI - PMC - PubMed
-
- CDC . 2021. Information for Healthcare Providers about Multisystem Inflammatory Syndrome in Children (MIS-C)https://www.cdc.gov/mis/mis-c/hcp/index.html
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

