ATOH8 binds SMAD3 to induce cellular senescence and prevent Ras-driven malignant transformation
- PMID: 36626550
- PMCID: PMC9934021
- DOI: 10.1073/pnas.2208927120
ATOH8 binds SMAD3 to induce cellular senescence and prevent Ras-driven malignant transformation
Abstract
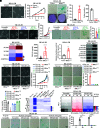
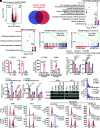
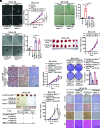
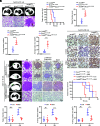
The process of oncogene-induced senescence (OIS) and the conversion between OIS and malignant transformation during carcinogenesis is poorly understood. Here, we show that following overactivation of oncogene Ras in lung epithelial cells, high-level transforming growth factor β1 (TGF-β1)-activated SMAD3, but not SMAD2 or SMAD4, plays a determinant role in inducing cellular senescence independent of the p53/p16/p15 senescence pathways. Importantly, SMAD3 binds a potential tumor suppressor ATOH8 to form a transcriptional complex that directly represses a series of cell cycle-promoting genes and consequently causes senescence in lung epithelial cells. Interestingly, the prosenescent SMAD3 converts to being oncogenic and essentially facilitates oncogenic Ras-driven malignant transformation. Furthermore, depleting Atoh8 rapidly accelerates oncogenic Ras-driven lung tumorigenesis, and lung cancers driven by mutant Ras and Atoh8 loss, but not by mutant Ras only, are sensitive to treatment of a specific SMAD3 inhibitor. Moreover, hypermethylation of the ATOH8 gene can be found in approximately 12% of clinical lung cancer cases. Together, our findings demonstrate not only epithelial cellular senescence directed by a potential tumor suppressor-controlled transcriptional program but also an important interplay between the prosenescent and transforming effects of TGF-β/SMAD3, potentially laying a foundation for developing early detection and anticancer strategies.
Keywords: Ras; SMAD3; TGF-β; cellular senescence; malignant transformation.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
TGF-beta signaling engages an ATM-CHK2-p53-independent RAS-induced senescence and prevents malignant transformation in human mammary epithelial cells.Proc Natl Acad Sci U S A. 2011 May 24;108(21):8668-73. doi: 10.1073/pnas.1015022108. Epub 2011 May 9. Proc Natl Acad Sci U S A. 2011. PMID: 21555587 Free PMC article.
-
Oncogenic ras represses transforming growth factor-beta /Smad signaling by degrading tumor suppressor Smad4.J Biol Chem. 2001 Aug 3;276(31):29531-7. doi: 10.1074/jbc.M100069200. Epub 2001 May 22. J Biol Chem. 2001. PMID: 11371552
-
Smad3 protein levels are modulated by Ras activity and during the cell cycle to dictate transforming growth factor-beta responses.J Biol Chem. 2010 Feb 26;285(9):6489-97. doi: 10.1074/jbc.M109.043877. Epub 2009 Dec 26. J Biol Chem. 2010. PMID: 20037158 Free PMC article.
-
Roles of Smad3 in TGF-beta signaling during carcinogenesis.Crit Rev Eukaryot Gene Expr. 2007;17(4):281-93. doi: 10.1615/critreveukargeneexpr.v17.i4.30. Crit Rev Eukaryot Gene Expr. 2007. PMID: 17725494 Free PMC article. Review.
-
A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling.J Cell Biochem. 2007 May 1;101(1):9-33. doi: 10.1002/jcb.21255. J Cell Biochem. 2007. PMID: 17340614 Review.
Cited by
-
Cellular senescence in cancer: molecular mechanisms and therapeutic _targets.MedComm (2020). 2024 Apr 24;5(5):e542. doi: 10.1002/mco2.542. eCollection 2024 May. MedComm (2020). 2024. PMID: 38660685 Free PMC article. Review.
-
Long-term severe hypoxia adaptation induces non-canonical EMT and a novel Wilms Tumor 1 (WT1) isoform.Cancer Gene Ther. 2024 Aug;31(8):1237-1250. doi: 10.1038/s41417-024-00795-3. Epub 2024 Jul 8. Cancer Gene Ther. 2024. PMID: 38977895 Free PMC article.
-
SMAD Proteins in TGF-β Signalling Pathway in Cancer: Regulatory Mechanisms and Clinical Applications.Diagnostics (Basel). 2023 Aug 26;13(17):2769. doi: 10.3390/diagnostics13172769. Diagnostics (Basel). 2023. PMID: 37685308 Free PMC article. Review.
References
-
- Li S., Balmain A., Counter C. M., A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat. Rev. Cancer 18, 767–777 (2018). - PubMed
-
- Johnson L., et al. , Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001). - PubMed
-
- Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W., Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997). - PubMed
-
- Stephen A. G., Esposito D., Bagni R. K., McCormick F., Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014). - PubMed
-
- Hernandez-Segura A., Nehme J., Demaria M., Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

