This is a preprint.
Single-cell multi-omic topic embedding reveals cell-type-specific and COVID-19 severity-related immune signatures
- PMID: 36778483
- PMCID: PMC9915637
- DOI: 10.1101/2023.01.31.526312
Single-cell multi-omic topic embedding reveals cell-type-specific and COVID-19 severity-related immune signatures
Update in
-
Single-cell multi-omics topic embedding reveals cell-type-specific and COVID-19 severity-related immune signatures.Cell Rep Methods. 2023 Aug 18;3(8):100563. doi: 10.1016/j.crmeth.2023.100563. eCollection 2023 Aug 28. Cell Rep Methods. 2023. PMID: 37671028 Free PMC article.
Abstract
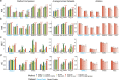
The advent of single-cell multi-omics sequencing technology makes it possible for re-searchers to leverage multiple modalities for individual cells and explore cell heterogeneity. However, the high dimensional, discrete, and sparse nature of the data make the downstream analysis particularly challenging. Most of the existing computational methods for single-cell data analysis are either limited to single modality or lack flexibility and interpretability. In this study, we propose an interpretable deep learning method called multi-omic embedded topic model (moETM) to effectively perform integrative analysis of high-dimensional single-cell multimodal data. moETM integrates multiple omics data via a product-of-experts in the encoder for efficient variational inference and then employs multiple linear decoders to learn the multi-omic signatures of the gene regulatory programs. Through comprehensive experiments on public single-cell transcriptome and chromatin accessibility data (i.e., scRNA+scATAC), as well as scRNA and proteomic data (i.e., CITE-seq), moETM demonstrates superior performance compared with six state-of-the-art single-cell data analysis methods on seven publicly available datasets. By applying moETM to the scRNA+scATAC data in human bone marrow mononuclear cells (BMMCs), we identified sequence motifs corresponding to the transcription factors that regulate immune gene signatures. Applying moETM analysis to CITE-seq data from the COVID-19 patients revealed not only known immune cell-type-specific signatures but also composite multi-omic biomarkers of critical conditions due to COVID-19, thus providing insights from both biological and clinical perspectives.
Conflict of interest statement
Declaration of interests
The authors declare no competing interests.
Figures





Similar articles
-
Single-cell multi-omics topic embedding reveals cell-type-specific and COVID-19 severity-related immune signatures.Cell Rep Methods. 2023 Aug 18;3(8):100563. doi: 10.1016/j.crmeth.2023.100563. eCollection 2023 Aug 28. Cell Rep Methods. 2023. PMID: 37671028 Free PMC article.
-
Protocol to perform integrative analysis of high-dimensional single-cell multimodal data using an interpretable deep learning technique.STAR Protoc. 2024 Jun 21;5(2):103066. doi: 10.1016/j.xpro.2024.103066. Epub 2024 May 14. STAR Protoc. 2024. PMID: 38748882 Free PMC article.
-
DEMOC: a deep embedded multi-omics learning approach for clustering single-cell CITE-seq data.Brief Bioinform. 2022 Sep 20;23(5):bbac347. doi: 10.1093/bib/bbac347. Brief Bioinform. 2022. PMID: 36047285
-
Profiling Chromatin Accessibility at Single-cell Resolution.Genomics Proteomics Bioinformatics. 2021 Apr;19(2):172-190. doi: 10.1016/j.gpb.2020.06.010. Epub 2021 Feb 11. Genomics Proteomics Bioinformatics. 2021. PMID: 33581341 Free PMC article. Review.
-
Multi-omic single cell sequencing: Overview and opportunities for kidney disease therapeutic development.Front Mol Biosci. 2023 Apr 5;10:1176856. doi: 10.3389/fmolb.2023.1176856. eCollection 2023. Front Mol Biosci. 2023. PMID: 37091871 Free PMC article. Review.
References
-
- Argelaguet R., Cuomo A. S., Stegle O. & Marioni J. C. Computational principles and challenges in single-cell data integration. Nature biotechnology 39, 1202–1215 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
