Chasing the mechanisms of ecologically adaptive salinity tolerance
- PMID: 36883005
- PMCID: PMC10721451
- DOI: 10.1016/j.xplc.2023.100571
Chasing the mechanisms of ecologically adaptive salinity tolerance
Abstract
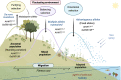
Plants adapted to challenging environments offer fascinating models of evolutionary change. Importantly, they also give information to meet our pressing need to develop resilient, low-input crops. With mounting environmental fluctuation-including temperature, rainfall, and soil salinity and degradation-this is more urgent than ever. Happily, solutions are hiding in plain sight: the adaptive mechanisms from natural adapted populations, once understood, can then be leveraged. Much recent insight has come from the study of salinity, a widespread factor limiting productivity, with estimates of 20% of all cultivated lands affected. This is an expanding problem, given increasing climate volatility, rising sea levels, and poor irrigation practices. We therefore highlight recent benchmark studies of ecologically adaptive salt tolerance in plants, assessing macro- and microevolutionary mechanisms, and the recently recognized role of ploidy and the microbiome on salinity adaptation. We synthesize insight specifically on naturally evolved adaptive salt-tolerance mechanisms, as these works move substantially beyond traditional mutant or knockout studies, to show how evolution can nimbly "tweak" plant physiology to optimize function. We then point to future directions to advance this field that intersect evolutionary biology, abiotic-stress tolerance, breeding, and molecular plant physiology.
Keywords: adaptation; ecology; evolution; microbiome; polyploidy; salinity.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Adaptive Mechanisms of Halophytes and Their Potential in Improving Salinity Tolerance in Plants.Int J Mol Sci. 2021 Oct 3;22(19):10733. doi: 10.3390/ijms221910733. Int J Mol Sci. 2021. PMID: 34639074 Free PMC article. Review.
-
Fluctuating selection on migrant adaptive sodium transporter alleles in coastal Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12443-E12452. doi: 10.1073/pnas.1816964115. Epub 2018 Dec 7. Proc Natl Acad Sci U S A. 2018. PMID: 30530653 Free PMC article.
-
Rewilding staple crops for the lost halophytism: Toward sustainability and profitability of agricultural production systems.Mol Plant. 2022 Jan 3;15(1):45-64. doi: 10.1016/j.molp.2021.12.003. Epub 2021 Dec 13. Mol Plant. 2022. PMID: 34915209 Review.
-
Engineering salinity tolerance in plants: progress and prospects.Planta. 2020 Mar 9;251(4):76. doi: 10.1007/s00425-020-03366-6. Planta. 2020. PMID: 32152761 Review.
-
Insights into the Transcriptomics of Crop Wild Relatives to Unravel the Salinity Stress Adaptive Mechanisms.Int J Mol Sci. 2023 Jun 6;24(12):9813. doi: 10.3390/ijms24129813. Int J Mol Sci. 2023. PMID: 37372961 Free PMC article. Review.
Cited by
-
Unveiling the influence of salinity on bacterial microbiome assembly of halophytes and crops.Environ Microbiome. 2024 Jul 18;19(1):49. doi: 10.1186/s40793-024-00592-3. Environ Microbiome. 2024. PMID: 39026296 Free PMC article.
-
Local cryptic diversity in salinity adaptation mechanisms in the wild outcrossing Brassica fruticulosa.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2407821121. doi: 10.1073/pnas.2407821121. Epub 2024 Sep 24. Proc Natl Acad Sci U S A. 2024. PMID: 39316046 Free PMC article.
-
Endophytic fungi are able to induce tolerance to salt stress in date palm seedlings (Phoenix dactylifera L.).Braz J Microbiol. 2024 Mar;55(1):759-775. doi: 10.1007/s42770-023-01216-7. Epub 2023 Dec 29. Braz J Microbiol. 2024. PMID: 38157149 Free PMC article.
References
-
- Abobatta W.F. Plants: Signaling Networks and Adaptive Mechanisms. Springer Cham; 2020. Plant responses and tolerance to extreme salinity: learning from halophyte tolerance to extreme salinity. Salt and Drought Stress Tolerance; pp. 177–210.
-
- Akyol T.Y., Sato S., Turkan I. Deploying root microbiome of halophytes to improve salinity tolerance of crops. Plant Biotechnol. Rep. 2020;14:143–150.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

