Dynamics of Chromatin Accessibility During Hematopoietic Stem Cell Differentiation Into Progressively Lineage-Committed Progeny
- PMID: 36945732
- PMCID: PMC10183972
- DOI: 10.1093/stmcls/sxad022
Dynamics of Chromatin Accessibility During Hematopoietic Stem Cell Differentiation Into Progressively Lineage-Committed Progeny
Abstract
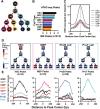
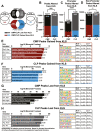
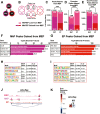
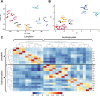
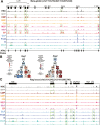
Epigenetic mechanisms regulate the multilineage differentiation capacity of hematopoietic stem cells (HSCs) into a variety of blood and immune cells. Mapping the chromatin dynamics of functionally defined cell populations will shed mechanistic insight into 2 major, unanswered questions in stem cell biology: how does epigenetic identity contribute to a cell type's lineage potential, and how do cascades of chromatin remodeling dictate ensuing fate decisions? Our recent work revealed evidence of multilineage gene priming in HSCs, where open cis-regulatory elements (CREs) exclusively shared between HSCs and unipotent lineage cells were enriched for DNA binding motifs of known lineage-specific transcription factors. Oligopotent progenitor populations operating between the HSCs and unipotent cells play essential roles in effecting hematopoietic homeostasis. To test the hypothesis that selective HSC-primed lineage-specific CREs remain accessible throughout differentiation, we used ATAC-seq to map the temporal dynamics of chromatin remodeling during progenitor differentiation. We observed epigenetic-driven clustering of oligopotent and unipotent progenitors into distinct erythromyeloid and lymphoid branches, with multipotent HSCs and MPPs associating with the erythromyeloid lineage. We mapped the dynamics of lineage-primed CREs throughout hematopoiesis and identified both unique and shared CREs as potential lineage reinforcement mechanisms at fate branch points. Additionally, quantification of genome-wide peak count and size revealed overall greater chromatin accessibility in HSCs, allowing us to identify HSC-unique peaks as putative regulators of self-renewal and multilineage potential. Finally, CRISPRi-mediated _targeting of ATACseq-identified putative CREs in HSCs allowed us to demonstrate the functional role of selective CREs in lineage-specific gene expression. These findings provide insight into the regulation of stem cell multipotency and lineage commitment throughout hematopoiesis and serve as a resource to test functional drivers of hematopoietic lineage fate.
Keywords: cell fate decisions; chromatin accessibility; epigenetics; hematopoiesis; hematopoietic stem and progenitor cells.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Conflict of interest statement
S.C. is a consultant for NextRNA Therapeutics and received honoraria for NextRNA from the American Association of Immunologists. All the other authors declared no potential conflicts of interest.
Figures


Similar articles
-
Chromatin accessibility maps provide evidence of multilineage gene priming in hematopoietic stem cells.Epigenetics Chromatin. 2021 Jan 6;14(1):2. doi: 10.1186/s13072-020-00377-1. Epigenetics Chromatin. 2021. PMID: 33407811 Free PMC article.
-
Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Human Developmental Hematopoiesis.Cell Stem Cell. 2021 Mar 4;28(3):472-487.e7. doi: 10.1016/j.stem.2020.11.015. Epub 2020 Dec 21. Cell Stem Cell. 2021. PMID: 33352111 Free PMC article.
-
Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells.Nature. 2018 Feb 1;554(7690):106-111. doi: 10.1038/nature25455. Epub 2018 Jan 3. Nature. 2018. PMID: 29298288
-
Hematopoietic stem cell lineage specification.Curr Opin Hematol. 2016 Jul;23(4):311-7. doi: 10.1097/MOH.0000000000000260. Curr Opin Hematol. 2016. PMID: 27135980 Review.
-
Differentiation-based model of hematopoietic stem cell functions and lineage pathways.Blood. 2018 Sep 13;132(11):1106-1113. doi: 10.1182/blood-2018-03-791517. Epub 2018 Jul 24. Blood. 2018. PMID: 30042097 Free PMC article. Review.
Cited by
-
HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo.bioRxiv [Preprint]. 2024 Jun 12:2024.06.10.598329. doi: 10.1101/2024.06.10.598329. bioRxiv. 2024. Update in: BMC Genomics. 2024 Dec 24;25(1):1240. doi: 10.1186/s12864-024-11162-9 PMID: 38915578 Free PMC article. Updated. Preprint.
-
Step by step analysis on gene datasets of growth phases in hematopoietic stem cells.Biochem Biophys Rep. 2024 May 30;39:101737. doi: 10.1016/j.bbrep.2024.101737. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 38881758 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

