The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression
- PMID: 36949076
- PMCID: PMC10033678
- DOI: 10.1038/s41467-023-37307-0
The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression
Abstract
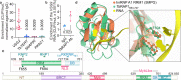
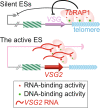
Trypanosoma brucei is a protozoan parasite that causes human African trypanosomiasis. Its major surface antigen VSG is expressed from subtelomeric loci in a strictly monoallelic manner. We previously showed that the telomere protein TbRAP1 binds dsDNA through its 737RKRRR741 patch to silence VSGs globally. How TbRAP1 permits expression of the single active VSG is unknown. Through NMR structural analysis, we unexpectedly identify an RNA Recognition Motif (RRM) in TbRAP1, which is unprecedented for RAP1 homologs. Assisted by the 737RKRRR741 patch, TbRAP1 RRM recognizes consensus sequences of VSG 3'UTRs in vitro and binds the active VSG RNA in vivo. Mutating conserved RRM residues abolishes the RNA binding activity, significantly decreases the active VSG RNA level, and derepresses silent VSGs. The competition between TbRAP1's RNA and dsDNA binding activities suggests a VSG monoallelic expression mechanism in which the active VSG's abundant RNA antagonizes TbRAP1's silencing effect, thereby sustaining its full-level expression.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
TbRAP1 has an unusual duplex DNA binding activity required for its telomere localization and VSG silencing.Sci Adv. 2020 Sep 18;6(38):eabc4065. doi: 10.1126/sciadv.abc4065. Print 2020 Sep. Sci Adv. 2020. PMID: 32948591 Free PMC article.
-
Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions.mSphere. 2020 Feb 26;5(1):e00027-20. doi: 10.1128/mSphere.00027-20. mSphere. 2020. PMID: 32102938 Free PMC article.
-
Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids.Nucleic Acids Res. 2017 Jun 2;45(10):5785-5796. doi: 10.1093/nar/gkx184. Nucleic Acids Res. 2017. PMID: 28334836 Free PMC article.
-
Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities.Pathogens. 2021 Jul 30;10(8):967. doi: 10.3390/pathogens10080967. Pathogens. 2021. PMID: 34451431 Free PMC article. Review.
-
Telomere and Subtelomere R-loops and Antigenic Variation in Trypanosomes.J Mol Biol. 2020 Jul 10;432(15):4167-4185. doi: 10.1016/j.jmb.2019.10.025. Epub 2019 Nov 2. J Mol Biol. 2020. PMID: 31682833 Free PMC article. Review.
Cited by
-
PI(3,4,5)P3 allosteric regulation of repressor activator protein 1 controls antigenic variation in trypanosomes.Elife. 2023 Nov 29;12:RP89331. doi: 10.7554/eLife.89331. Elife. 2023. PMID: 38019264 Free PMC article.
-
Telomere maintenance in African trypanosomes.Front Mol Biosci. 2023 Nov 24;10:1302557. doi: 10.3389/fmolb.2023.1302557. eCollection 2023. Front Mol Biosci. 2023. PMID: 38074093 Free PMC article. Review.
-
An allele-selective inter-chromosomal protein bridge supports monogenic antigen expression in the African trypanosome.Nat Commun. 2023 Dec 11;14(1):8200. doi: 10.1038/s41467-023-44043-y. Nat Commun. 2023. PMID: 38081826 Free PMC article.
-
Unwrap RAP1's Mystery at Kinetoplastid Telomeres.Biomolecules. 2024 Jan 4;14(1):67. doi: 10.3390/biom14010067. Biomolecules. 2024. PMID: 38254667 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

