Blood-Based Proteomic Profiling Identifies Potential Biomarker Candidates and Pathogenic Pathways in Dementia
- PMID: 37175824
- PMCID: PMC10179172
- DOI: 10.3390/ijms24098117
Blood-Based Proteomic Profiling Identifies Potential Biomarker Candidates and Pathogenic Pathways in Dementia
Abstract
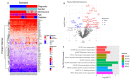
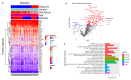
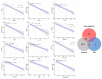
Dementia is a progressive and debilitating neurological disease that affects millions of people worldwide. Identifying the minimally invasive biomarkers associated with dementia that could provide insights into the disease pathogenesis, improve early diagnosis, and facilitate the development of effective treatments is pressing. Proteomic studies have emerged as a promising approach for identifying the protein biomarkers associated with dementia. This pilot study aimed to investigate the plasma proteome profile and identify a panel of various protein biomarkers for dementia. We used a high-throughput proximity extension immunoassay to quantify 1090 proteins in 122 participants (22 with dementia, 64 with mild cognitive impairment (MCI), and 36 controls with normal cognitive function). Limma-based differential expression analysis reported the dysregulation of 61 proteins in the plasma of those with dementia compared with controls, and machine learning algorithms identified 17 stable diagnostic biomarkers that differentiated individuals with AUC = 0.98 ± 0.02. There was also the dysregulation of 153 plasma proteins in individuals with dementia compared with those with MCI, and machine learning algorithms identified 8 biomarkers that classified dementia from MCI with an AUC of 0.87 ± 0.07. Moreover, multiple proteins selected in both diagnostic panels such as NEFL, IL17D, WNT9A, and PGF were negatively correlated with cognitive performance, with a correlation coefficient (r2) ≤ -0.47. Gene Ontology (GO) and pathway analysis of dementia-associated proteins implicated immune response, vascular injury, and extracellular matrix organization pathways in dementia pathogenesis. In conclusion, the combination of high-throughput proteomics and machine learning enabled us to identify a blood-based protein signature capable of potentially differentiating dementia from MCI and cognitively normal controls. Further research is required to validate these biomarkers and investigate the potential underlying mechanisms for the development of dementia.
Keywords: MCI; Olink assay; biomarkers; dementia; machine learning; plasma proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Neuroinflammation and Alzheimer's Disease: A Machine Learning Approach to CSF Proteomics.Cells. 2021 Jul 29;10(8):1930. doi: 10.3390/cells10081930. Cells. 2021. PMID: 34440700 Free PMC article.
-
A Community-Based Study Identifying Metabolic Biomarkers of Mild Cognitive Impairment and Alzheimer's Disease Using Artificial Intelligence and Machine Learning.J Alzheimers Dis. 2020;78(4):1381-1392. doi: 10.3233/JAD-200305. J Alzheimers Dis. 2020. PMID: 33164929
-
Blood phosphorylated tau 181 as a biomarker for Alzheimer's disease: a diagnostic performance and prediction modelling study using data from four prospective cohorts.Lancet Neurol. 2020 May;19(5):422-433. doi: 10.1016/S1474-4422(20)30071-5. Lancet Neurol. 2020. PMID: 32333900
-
Cerebrospinal fluid proteomics and biological heterogeneity in Alzheimer's disease: A literature review.Crit Rev Clin Lab Sci. 2020 Mar;57(2):86-98. doi: 10.1080/10408363.2019.1670613. Epub 2019 Nov 7. Crit Rev Clin Lab Sci. 2020. PMID: 31694431 Review.
-
Machine learning models of plasma proteomic data predict mood in chronic stroke and tie it to aberrant peripheral immune responses.Brain Behav Immun. 2023 Nov;114:144-153. doi: 10.1016/j.bbi.2023.08.002. Epub 2023 Aug 7. Brain Behav Immun. 2023. PMID: 37557961 Free PMC article. Review.
Cited by
-
Alzheimer's disease early diagnostic and staging biomarkers revealed by large-scale cerebrospinal fluid and serum proteomic profiling.Innovation (Camb). 2024 Jan 2;5(1):100544. doi: 10.1016/j.xinn.2023.100544. eCollection 2024 Jan 8. Innovation (Camb). 2024. PMID: 38235188 Free PMC article.
-
Molecular Mechanisms Underlying Chronic and Degenerative Diseases.Int J Mol Sci. 2023 Aug 7;24(15):12507. doi: 10.3390/ijms241512507. Int J Mol Sci. 2023. PMID: 37569882 Free PMC article.
-
Biological gases, oxidative stress, artificial intelligence, and machine learning for neurodegeneration and metabolic disorders.Med Gas Res. 2025 Mar 1;15(1):145-147. doi: 10.4103/mgr.MEDGASRES-D-24-00059. Epub 2024 Oct 2. Med Gas Res. 2025. PMID: 39436188 Free PMC article. No abstract available.
-
Plasma proteomics and lipidomics facilitate elucidation of the link between Alzheimer's disease development and vessel wall fragility.Sci Rep. 2024 Aug 27;14(1):19901. doi: 10.1038/s41598-024-71097-9. Sci Rep. 2024. PMID: 39191863 Free PMC article.
References
-
- Alzheimer’s Association 2018 Alzheimer’s Disease Facts and Figures. Alzheimer’s Dement. 2018;14:367–429. doi: 10.1016/j.jalz.2018.02.001. - DOI
-
- Barker W.W., Luis C.A., Kashuba A., Luis M., Harwood D.G., Loewenstein D., Waters C., Jimison P., Shepherd E., Sevush S., et al. Relative Frequencies of Alzheimer Disease, Lewy Body, Vascular and Frontotemporal Dementia, and Hippocampal Sclerosis in the State of Florida Brain Bank. Alzheimer Dis. Assoc. Disord. 2002;16:203–212. doi: 10.1097/00002093-200210000-00001. - DOI - PubMed
-
- Prince M.J., Wimo A., Guerchet M.M., Ali G.C., Wu Y.T., Prina M. World Alzheimer Report 2015: The Global Impact of Dementia. Alzheimer’s Disease International; London, UK: 2015.
-
- United Nations Department of Economic and Social Affairs Population Division . World Population Ageing 2019. Volume Highlights. United Nations Department of Economic and Social Affairs Population Division; New York, NY, USA: 2019.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

