Regulation of Flowering Time and Other Developmental Plasticities by 3' Splicing Factor-Mediated Alternative Splicing in Arabidopsis thaliana
- PMID: 37836248
- PMCID: PMC10575287
- DOI: 10.3390/plants12193508
Regulation of Flowering Time and Other Developmental Plasticities by 3' Splicing Factor-Mediated Alternative Splicing in Arabidopsis thaliana
Abstract
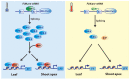
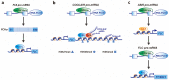
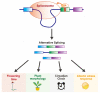
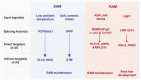
Plants, as sessile organisms, show a high degree of plasticity in their growth and development and have various strategies to cope with these alterations under continuously changing environments and unfavorable stress conditions. In particular, the floral transition from the vegetative and reproductive phases in the shoot apical meristem (SAM) is one of the most important developmental changes in plants. In addition, meristem regions, such as the SAM and root apical meristem (RAM), which continually generate new lateral organs throughout the plant life cycle, are important sites for developmental plasticity. Recent findings have shown that the prevailing type of alternative splicing (AS) in plants is intron retention (IR) unlike in animals; thus, AS is an important regulatory mechanism conferring plasticity for plant growth and development under various environmental conditions. Although eukaryotes exhibit some similarities in the composition and dynamics of their splicing machinery, plants have differences in the 3' splicing characteristics governing AS. Here, we summarize recent findings on the roles of 3' splicing factors and their interacting partners in regulating the flowering time and other developmental plasticities in Arabidopsis thaliana.
Keywords: 3’ splicing factors; alternative splicing; developmental plasticity; flowering time; root apical meristem; shoot apical meristem.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
SKI-INTERACTING PROTEIN interacts with SHOOT MERISTEMLESS to regulate shoot apical meristem formation.Plant Physiol. 2022 Aug 1;189(4):2193-2209. doi: 10.1093/plphys/kiac241. Plant Physiol. 2022. PMID: 35640153 Free PMC article.
-
A gibberellin methyltransferase modulates the timing of floral transition at the Arabidopsis shoot meristem.Physiol Plant. 2020 Dec;170(4):474-487. doi: 10.1111/ppl.13146. Epub 2020 Jun 16. Physiol Plant. 2020. PMID: 32483836
-
XAANTAL2 (AGL14) Is an Important Component of the Complex Gene Regulatory Network that Underlies Arabidopsis Shoot Apical Meristem Transitions.Mol Plant. 2015 May;8(5):796-813. doi: 10.1016/j.molp.2015.01.017. Epub 2015 Jan 28. Mol Plant. 2015. PMID: 25636918
-
The vascular plants: open system of growth.Dev Genes Evol. 2017 Mar;227(2):129-157. doi: 10.1007/s00427-016-0572-1. Epub 2017 Feb 18. Dev Genes Evol. 2017. PMID: 28214944 Review.
-
The Roles of Plant Hormones and Their Interactions with Regulatory Genes in Determining Meristem Activity.Int J Mol Sci. 2019 Aug 20;20(16):4065. doi: 10.3390/ijms20164065. Int J Mol Sci. 2019. PMID: 31434317 Free PMC article. Review.
References
-
- Xie X., Wu N. Introns in higher plant genes. Chin. Sci. Bull. 2002;47:1409–1415. doi: 10.1360/02tb9311. - DOI
Publication types
Grants and funding
- 2021R1I1A3050195/the Basic Science Research Program of the National Research Foundation (NRF) funded by the Ministry of Education of the Republic of Korea
- 2021R1F1A1047817/the Basic Science Research Program of the National Research Foundation (NRF) funded by the Ministry of Education of the Republic of Korea
- No/Korea University
LinkOut - more resources
Full Text Sources
Research Materials

