Plasma Proteomics to Identify Drug _targets for Ischemic Heart Disease
- PMID: 37940228
- PMCID: PMC10641761
- DOI: 10.1016/j.jacc.2023.09.804
Plasma Proteomics to Identify Drug _targets for Ischemic Heart Disease
Abstract
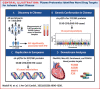
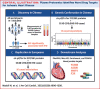
Background: Integrated analyses of plasma proteomic and genetic markers in prospective studies can clarify the causal relevance of proteins and discover novel _targets for ischemic heart disease (IHD) and other diseases.
Objectives: The purpose of this study was to examine associations of proteomics and genetics data with IHD in population studies to discover novel preventive treatments.
Methods: We conducted a nested case-cohort study in the China Kadoorie Biobank (CKB) involving 1,971 incident IHD cases and 2,001 subcohort participants who were genotyped and free of prior cardiovascular disease. We measured 1,463 proteins in the stored baseline samples using the OLINK EXPLORE panel. Cox regression yielded adjusted HRs for IHD associated with individual proteins after accounting for multiple testing. Moreover, cis-protein quantitative loci (pQTLs) identified for proteins in genome-wide association studies of CKB and of UK Biobank were used as instrumental variables in separate 2-sample Mendelian randomization (MR) studies involving global CARDIOGRAM+C4D consortium (210,842 IHD cases and 1,378,170 controls).
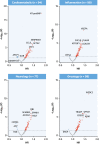
Results: Overall 361 proteins were significantly associated at false discovery rate <0.05 with risk of IHD (349 positively, 12 inversely) in CKB, including N-terminal prohormone of brain natriuretic peptide and proprotein convertase subtilisin/kexin type 9. Of these 361 proteins, 212 had cis-pQTLs in CKB, and MR analyses of 198 variants in CARDIOGRAM+C4D identified 13 proteins that showed potentially causal associations with IHD. Independent MR analyses of 307 cis-pQTLs identified in Europeans replicated associations for 4 proteins (FURIN, proteinase-activated receptor-1, Asialoglycoprotein receptor-1, and matrix metalloproteinase-3). Further downstream analyses showed that FURIN, which is highly expressed in endothelial cells, is a potential novel _target and matrix metalloproteinase-3 a potential repurposing _target for IHD.
Conclusions: Integrated analyses of proteomic and genetic data in Chinese and European adults provided causal support for FURIN and multiple other proteins as potential novel drug _targets for treatment of IHD.
Keywords: Mendelian randomization; diverse populations; drug _target; genetics; ischemic heart disease; plasma proteomics; prospective studies.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures The CKB baseline survey and the first resurvey were supported by the Kadoorie Charitable Foundation in Hong Kong. The long-term follow-up and subsequent resurveys have been supported by Wellcome grants to Oxford University (212946/Z/18/Z, 202922/Z/16/Z, 104085/Z/14/Z, 088158/Z/09/Z) and grants from the National Natural Science Foundation of China (82192901, 82192904, 82192900) and from the National Key Research and Development Program of China (2016YFC0900500). The UK Medical Research Council (MC_UU_00017/1, MC_UU_12026/2, MC_U137686851), Cancer Research UK (C16077/A29186, C500/A16896), and British Heart Foundation (CH/1996001/9454) provide core funding to the Clinical Trial Service Unit and Epidemiological Studies Unit, Oxford University for the project. The proteomic assays were supported by a BHF Intermediate Clinical Research Fellowship to MVH (FS/18/23/33512), Novo Nordisk, and OLINK. DNA extraction and genotyping were supported by GlaxoSmithKline and the UK Medical Research Council (MC-PC-13049, MC-PC-14135). Dr Holmes is currently employed by 23andMe (and owns stock in 23andMe, Inc). Dr Howson is a full-time employee of Novo Nordisk Research Centre Oxford Limited and owns shares in Novo Nordisk. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures




Comment in
-
Multiomics Insights to Accelerate Drug Development: Will They Hold Their Promises?J Am Coll Cardiol. 2023 Nov 14;82(20):1932-1935. doi: 10.1016/j.jacc.2023.09.801. J Am Coll Cardiol. 2023. PMID: 37940230 No abstract available.
Similar articles
-
Effect of n-3 polyunsaturated fatty acids on ischemic heart disease and cardiometabolic risk factors: a two-sample Mendelian randomization study.BMC Cardiovasc Disord. 2021 Nov 8;21(1):532. doi: 10.1186/s12872-021-02342-6. BMC Cardiovasc Disord. 2021. PMID: 34749668 Free PMC article. Clinical Trial.
-
Mendelian randomization and genetic colocalization infer the effects of the multi-tissue proteome on 211 complex disease-related phenotypes.Genome Med. 2022 Dec 12;14(1):140. doi: 10.1186/s13073-022-01140-9. Genome Med. 2022. PMID: 36510323 Free PMC article.
-
Effect of glutamate and aspartate on ischemic heart disease, blood pressure, and diabetes: a Mendelian randomization study.Am J Clin Nutr. 2019 Apr 1;109(4):1197-1206. doi: 10.1093/ajcn/nqy362. Am J Clin Nutr. 2019. PMID: 30949673
-
PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses.J Am Coll Cardiol. 2010 Jun 22;55(25):2833-42. doi: 10.1016/j.jacc.2010.02.044. J Am Coll Cardiol. 2010. PMID: 20579540 Review.
-
Vegetarian and vegan diets and the risk of cardiovascular disease, ischemic heart disease and stroke: a systematic review and meta-analysis of prospective cohort studies.Eur J Nutr. 2023 Feb;62(1):51-69. doi: 10.1007/s00394-022-02942-8. Epub 2022 Aug 27. Eur J Nutr. 2023. PMID: 36030329 Free PMC article. Review.
Cited by
-
The Role of Furin and Its Therapeutic Potential in Cardiovascular Disease Risk.Int J Mol Sci. 2024 Aug 26;25(17):9237. doi: 10.3390/ijms25179237. Int J Mol Sci. 2024. PMID: 39273186 Free PMC article. Review.
-
Plasma proteomics-based organ-specific aging for all-cause mortality and cause-specific mortality: a prospective cohort study.Geroscience. 2024 Oct 31. doi: 10.1007/s11357-024-01411-w. Online ahead of print. Geroscience. 2024. PMID: 39477866
-
Cuproptosis and copper deficiency in ischemic vascular injury and repair.Apoptosis. 2024 Aug;29(7-8):1007-1018. doi: 10.1007/s10495-024-01969-y. Epub 2024 Apr 22. Apoptosis. 2024. PMID: 38649508 Review.
-
The serum soluble ASGR1 concentration is elevated in patients with coronary artery disease and is associated with inflammatory markers.Lipids Health Dis. 2024 Mar 27;23(1):89. doi: 10.1186/s12944-024-02054-8. Lipids Health Dis. 2024. PMID: 38539180 Free PMC article.
-
Risk prediction of ischemic heart disease using plasma proteomics, conventional risk factors and polygenic scores in Chinese and European adults.Eur J Epidemiol. 2024 Nov;39(11):1229-1240. doi: 10.1007/s10654-024-01168-8. Epub 2024 Nov 22. Eur J Epidemiol. 2024. PMID: 39578299 Free PMC article.
References
-
- Williams S.A., Ostroff R., Hinterberg M.A., et al. A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci Transl Med. 2022;14 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

