UBE3C Facilitates the ER-Associated and Peripheral Degradation of Misfolded CFTR
- PMID: 38067172
- PMCID: PMC10706245
- DOI: 10.3390/cells12232741
UBE3C Facilitates the ER-Associated and Peripheral Degradation of Misfolded CFTR
Abstract
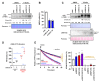
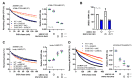
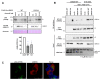
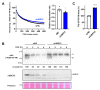
The ubiquitin E3 ligase UBE3C promotes the proteasomal degradation of cytosolic proteins and endoplasmic reticulum (ER) membrane proteins. UBE3C is proposed to function downstream of the RNF185/MBRL ER-associated degradation (ERAD) branch, contributing to the ERAD of select membrane proteins. Here, we report that UBE3C facilitates the ERAD of misfolded CFTR, even in the absence of both RNF185 and its functional ortholog RNF5 (RNF5/185). Unlike RNF5/185, UBE3C had a limited impact on the ubiquitination of misfolded CFTR. UBE3C knockdown (KD) resulted in an additional increase in the functional ∆F508-CFTR channels on the plasma membrane when combined with the RNF5/185 ablation, particularly in the presence of clinically used CFTR modulators. Interestingly, although UBE3C KD failed to attenuate the ERAD of insig-1, it reduced the ERAD of misfolded ∆Y490-ABCB1 and increased cell surface expression. UBE3C KD also stabilized the mature form of ∆F508-CFTR and increased the cell surface level of T70-CFTR, a class VI CFTR mutant. These results suggest that UBE3C plays a vital role in the ERAD of misfolded CFTR and ABCB1, even within the RNF5/185-independent ERAD pathway, and it may also be involved in maintaining the peripheral quality control of CFTR.
Keywords: ABCB1; CFTR; ERAD; RNF185; RNF5; UBE3C; protein quality control; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that _targets cystic fibrosis transmembrane conductance regulator (CFTR).J Biol Chem. 2013 Oct 25;288(43):31177-91. doi: 10.1074/jbc.M113.470500. Epub 2013 Sep 9. J Biol Chem. 2013. PMID: 24019521 Free PMC article.
-
HERC3 facilitates ERAD of select membrane proteins by recognizing membrane-spanning domains.J Cell Biol. 2024 Jul 1;223(7):e202308003. doi: 10.1083/jcb.202308003. Epub 2024 May 9. J Cell Biol. 2024. PMID: 38722278 Free PMC article.
-
Small-molecule correctors divert CFTR-F508del from ERAD by stabilizing sequential folding states.Mol Biol Cell. 2024 Feb 1;35(2):ar15. doi: 10.1091/mbc.E23-08-0336. Epub 2023 Nov 29. Mol Biol Cell. 2024. PMID: 38019608 Free PMC article.
-
How Is the Fidelity of Proteins Ensured in Terms of Both Quality and Quantity at the Endoplasmic Reticulum? Mechanistic Insights into E3 Ubiquitin Ligases.Int J Mol Sci. 2021 Feb 19;22(4):2078. doi: 10.3390/ijms22042078. Int J Mol Sci. 2021. PMID: 33669844 Free PMC article. Review.
-
Regulation of CFTR Biogenesis by the Proteostatic Network and Pharmacological Modulators.Int J Mol Sci. 2020 Jan 10;21(2):452. doi: 10.3390/ijms21020452. Int J Mol Sci. 2020. PMID: 31936842 Free PMC article. Review.
Cited by
-
_targeting ubiquitination machinery in cystic fibrosis: Where do we stand?Cell Mol Life Sci. 2024 Jun 18;81(1):271. doi: 10.1007/s00018-024-05295-z. Cell Mol Life Sci. 2024. PMID: 38888668 Free PMC article. Review.
-
Ligand-based virtual-screening identified a novel CFTR ligand which improves the defective cell surface expression of misfolded ABC transporters.Front Pharmacol. 2024 Apr 11;15:1370676. doi: 10.3389/fphar.2024.1370676. eCollection 2024. Front Pharmacol. 2024. PMID: 38666024 Free PMC article.
References
-
- Rich D.P., Anderson M.P., Gregory R.J., Cheng S.H., Paul S., Jefferson D.M., McCann J.D., Klinger K.W., Smith A.E., Welsh M.J. Expression of Cystic-Fibrosis Transmembrane Conductance Regulator Corrects Defective Chloride Channel Regulation in Cystic-Fibrosis Airway Epithelial-Cells. Nature. 1990;347:358–363. doi: 10.1038/347358a0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

