Cellular and molecular mechanisms of fibrosis and resolution in bleomycin-induced pulmonary fibrosis mouse model revealed by spatial transcriptome analysis
- PMID: 38125541
- PMCID: PMC10730595
- DOI: 10.1016/j.heliyon.2023.e22461
Cellular and molecular mechanisms of fibrosis and resolution in bleomycin-induced pulmonary fibrosis mouse model revealed by spatial transcriptome analysis
Abstract
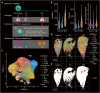
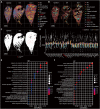
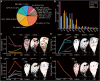
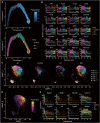
The bleomycin-induced pulmonary fibrosis mouse model is commonly used in idiopathic pulmonary fibrosis research, but its cellular and molecular changes and efficiency as a model at the molecular level are not fully understood. In this study, we used spatial transcriptome technology to investigate the cellular and molecular changes in the lungs of bleomycin-induced pulmonary fibrosis mouse models. Our analyses revealed cell dynamics during fibrosis in epithelial cells, mesenchymal cells, immunocytes, and erythrocytes with their spatial distribution available. We confirmed the differentiation of the alveolar type II (AT2) cell type expressing Krt8, and we inferred their trajectories from both the AT2 cells and club cells. In addition to the fibrosis process, we also noticed evidence of self-resolving, especially to identify possible self-resolving related genes, including Prkca. Our findings provide insights into the cellular and molecular mechanisms underlying fibrosis resolution and represent the first spatiotemporal transcriptome dataset of the bleomycin-induced fibrosis mouse model.
Keywords: Bleomycin-induced pulmonary fibrosis; Self-resolving; spatial transcriptome.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Glutamine Metabolism Is Required for Alveolar Regeneration during Lung Injury.Biomolecules. 2022 May 22;12(5):728. doi: 10.3390/biom12050728. Biomolecules. 2022. PMID: 35625656 Free PMC article.
-
The increase of microRNA-21 during lung fibrosis and its contribution to epithelial-mesenchymal transition in pulmonary epithelial cells.Respir Res. 2013 Sep 24;14(1):95. doi: 10.1186/1465-9921-14-95. Respir Res. 2013. PMID: 24063588 Free PMC article.
-
Mesenchymal stromal cells attenuate alveolar type 2 cells senescence through regulating NAMPT-mediated NAD metabolism.Stem Cell Res Ther. 2022 Jan 10;13(1):12. doi: 10.1186/s13287-021-02688-w. Stem Cell Res Ther. 2022. PMID: 35012648 Free PMC article.
-
Prominin-1/CD133+ lung epithelial progenitors protect from bleomycin-induced pulmonary fibrosis.Am J Respir Crit Care Med. 2009 May 15;179(10):939-49. doi: 10.1164/rccm.200809-1390OC. Epub 2009 Feb 20. Am J Respir Crit Care Med. 2009. PMID: 19234103
-
Induced pluripotent stem cell-derived lung alveolar epithelial type II cells reduce damage in bleomycin-induced lung fibrosis.Stem Cell Res Ther. 2020 Jun 3;11(1):213. doi: 10.1186/s13287-020-01726-3. Stem Cell Res Ther. 2020. PMID: 32493487 Free PMC article.
References
-
- Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S., Fernandez R., Akbarpour M., Chen C.I., Ren Z., et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. - DOI - PMC - PubMed
-
- Adams T.S., Schupp J.C., Poli S., Ayaub E.A., Neumark N., Ahangari F., Chu S.G., Raby B.A., DeIuliis G., Januszyk M., et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba1983. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

