Conversion of the CG specific M.MpeI DNA methyltransferase into an enzyme predominantly methylating CCA and CCC sites
- PMID: 38164970
- PMCID: PMC10899764
- DOI: 10.1093/nar/gkad1217
Conversion of the CG specific M.MpeI DNA methyltransferase into an enzyme predominantly methylating CCA and CCC sites
Abstract
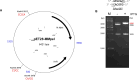
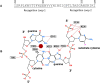
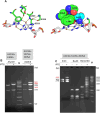
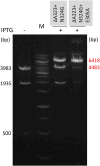
We used structure guided mutagenesis and directed enzyme evolution to alter the specificity of the CG specific bacterial DNA (cytosine-5) methyltransferase M.MpeI. Methylation specificity of the M.MpeI variants was characterized by digestions with methylation sensitive restriction enzymes and by measuring incorporation of tritiated methyl groups into double-stranded oligonucleotides containing single CC, CG, CA or CT sites. Site specific mutagenesis steps designed to disrupt the specific contacts between the enzyme and the non-substrate base pair of the _target sequence (5'-CG/5'-CG) yielded M.MpeI variants with varying levels of CG specific and increasing levels of CA and CC specific MTase activity. Subsequent random mutagenesis of the _target recognizing domain coupled with selection for non-CG specific methylation yielded a variant, which predominantly methylates CC dinucleotides, has very low activity on CG and CA sites, and no activity on CT sites. This M.MpeI variant contains a one amino acid deletion (ΔA323) and three substitutions (N324G, R326G and E305N) in the _target recognition domain. The mutant enzyme has very strong preference for A and C in the 3' flanking position making it a CCA and CCC specific DNA methyltransferase.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

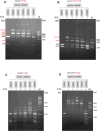
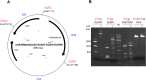


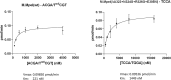
Similar articles
-
Circularly permuted variants of two CG-specific prokaryotic DNA methyltransferases.PLoS One. 2018 May 10;13(5):e0197232. doi: 10.1371/journal.pone.0197232. eCollection 2018. PLoS One. 2018. PMID: 29746549 Free PMC article.
-
Identification, expression, and purification of DNA cytosine 5-methyltransferases with short recognition sequences.BMC Biotechnol. 2022 Nov 4;22(1):33. doi: 10.1186/s12896-022-00765-3. BMC Biotechnol. 2022. PMID: 36333700 Free PMC article.
-
Altering the sequence specificity of HaeIII methyltransferase by directed evolution using in vitro compartmentalization.Protein Eng Des Sel. 2004 Jan;17(1):3-11. doi: 10.1093/protein/gzh001. Protein Eng Des Sel. 2004. PMID: 14985532
-
[DNA methyltransferases: the role in regulation of gene expression and biological processes].Yi Chuan. 2009 Nov;31(11):1087-93. doi: 10.3724/sp.j.1005.2009.01087. Yi Chuan. 2009. PMID: 19933088 Review. Chinese.
-
[DNA methyltransferases: classification, functions and research progress].Yi Chuan. 2009 Sep;31(9):903-12. doi: 10.3724/sp.j.1005.2009.00903. Yi Chuan. 2009. PMID: 19819843 Review. Chinese.
References
-
- Bochtler M., Fernandes H. DNA adenine methylation in eukaryotes: enzymatic mark or a form of DNA damage?. Bioessays. 2021; 43:2000243. - PubMed
-
- Jurkowska R.Z., and Jeltsch A. (2022) Mechanisms and biological roles of DNA methyltransferases and DNA methylation: from past achievements to future challenges. In: Jeltsch A., Jurkowska R.Z. (eds.) DNA Methyltr ansfer ases - Role and Function. Advances in Experimental Medicine and Biology. Vol. 1389, Springer, Cham, pp. 1–19. - PubMed
-
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005; 74:481–514. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

