Identification of Tissue miRNA Signatures for Pancreatic Ductal Adenocarcinoma
- PMID: 38398215
- PMCID: PMC10887387
- DOI: 10.3390/cancers16040824
Identification of Tissue miRNA Signatures for Pancreatic Ductal Adenocarcinoma
Abstract
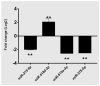
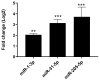
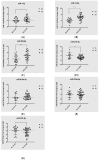
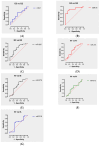
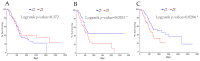
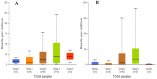
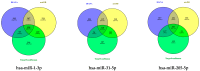
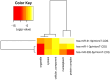
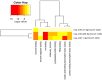
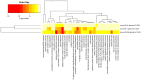
Pancreatic ductal adenocarcinoma (PDAC), a neoplasm of the gastrointestinal tract, is the most common pancreatic malignancy (90%) and the fourth highest cause of cancer mortality worldwide. Surgery intervention is currently the only strategy able to offer an advantage in terms of overall survival, but prognosis remains poor even for operated patients. Therefore, the development of robust biomarkers for early diagnosis and prognostic stratification in clinical practice is urgently needed. In this work, we investigated deregulated microRNAs (miRNAs) in tissues from PDAC patients with high (G3) or low (G2) histological grade and with (N+) or without (N-) lymph node metastases. miRNA expression profiling was performed by a comprehensive PCR array and subsequent validation by RT-qPCR. The results showed a significant increase in miR-1-3p, miR-31-5p, and miR-205-5p expression in G3 compared to G2 patients (** p < 0.01; *** p < 0.001; *** p < 0.001). miR-518d-3p upregulation and miR-215-5p downregulation were observed in N+ compared to N- patients. A statistical analysis performed using OncomiR program showed the significant involvement (p < 0.05) of two miRNAs (miR-31 and miR-205) in the histological grade of PDAC patients. Also, an expression analysis in PDAC patients showed that miR-31 and miR-205 had the highest expression at grade 3 compared with normal and other tumor grades. Overall, survival plots confirmed that the overexpression of miR-31 and miR-205 was significantly correlated with decreased survival in TCGA PDAC clinical samples. A KEGG pathway analysis showed that all three miRNAs are involved in the regulation of multiple pathways, including the Hippo signaling, adherens junction and microRNAs in cancer, along with several _target genes. Based on in silico analysis and experimental validation, our study suggests the potential role of miR-1-3p, miR-31-5p, and miR-205-5p as useful clinical biomarkers and putative therapeutic _targets in PDAC, which should be further investigated to determine the specific molecular processes affected by their aberrant expression.
Keywords: biomarkers; diagnosis; microRNAs; pancreatic ductal adenocarcinoma; prognosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Identification and validation of the microRNAs and hub genes for pancreatic ductal adenocarcinoma by an integrated bioinformatic analysis.J Gastrointest Oncol. 2023 Apr 29;14(2):719-732. doi: 10.21037/jgo-23-192. J Gastrointest Oncol. 2023. PMID: 37201049 Free PMC article.
-
Serum Exosomal miRNA-1226 as Potential Biomarker of Pancreatic Ductal Adenocarcinoma.Onco _targets Ther. 2021 Feb 26;14:1441-1451. doi: 10.2147/OTT.S296816. eCollection 2021. Onco _targets Ther. 2021. PMID: 33664577 Free PMC article.
-
MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma.Clin Cancer Res. 2012 Jan 15;18(2):534-45. doi: 10.1158/1078-0432.CCR-11-0679. Epub 2011 Nov 23. Clin Cancer Res. 2012. PMID: 22114136
-
Circulating microRNAs as Potential Diagnostic, Prognostic and Therapeutic _targets in Pancreatic Cancer.Curr Pharm Des. 2016;22(42):6444-6450. doi: 10.2174/1381612822666160817095047. Curr Pharm Des. 2016. PMID: 27539232 Review.
-
Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance.J Exp Clin Cancer Res. 2013 Sep 28;32(1):71. doi: 10.1186/1756-9966-32-71. J Exp Clin Cancer Res. 2013. PMID: 24289824 Free PMC article. Review.
Cited by
-
MicroRNAs in Pancreatic Cancer: Advances in Biomarker Discovery and Therapeutic Implications.Int J Mol Sci. 2024 Mar 31;25(7):3914. doi: 10.3390/ijms25073914. Int J Mol Sci. 2024. PMID: 38612727 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

