Unlocking Nature's Toolbox: glutamate-inducible recombinant protein production from the Komagatella phaffii PEPCK promoter
- PMID: 38402195
- PMCID: PMC10893637
- DOI: 10.1186/s12934-024-02340-1
Unlocking Nature's Toolbox: glutamate-inducible recombinant protein production from the Komagatella phaffii PEPCK promoter
Abstract
Background: Komagataella phaffii (a.k.a. Pichia pastoris) harbors a glutamate utilization pathway in which synthesis of glutamate dehydrogenase 2 and phosphoenolpyruvate carboxykinase (PEPCK) is induced by glutamate. Glutamate-inducible synthesis of these enzymes is regulated by Rtg1p, a cytosolic, basic helix-loop-helix protein. Here, we report food-grade monosodium glutamate (MSG)-inducible recombinant protein production from K. phaffii PEPCK promoter (PPEPCK) using green fluorescent protein (GFP) and receptor binding domain of SARS-CoV-2 virus (RBD) as model proteins.
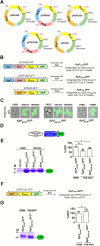
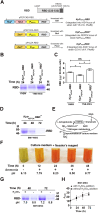
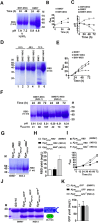
Results: PPEPCK-RBD/GFP expression cassette was integrated at two different sites in the genome to improve recombinant protein yield from PPEPCK. The traditional, methanol-inducible alcohol oxidase 1 promoter (PAOX1) was used as the benchmark. Initial studies carried out with MSG as the inducer resulted in low recombinant protein yield. A new strategy employing MSG/ethanol mixed feeding improved biomass generation as well as recombinant protein yield. Cell density of 100-120 A600 units/ml was achieved after 72 h of induction in shake flask cultivations, resulting in recombinant protein yield from PPEPCK that is comparable or even higher than that from PAOX1.
Conclusions: We have designed an induction medium for recombinant protein production from K. phaffii PPEPCK in shake flask cultivations. It consists of 1.0% yeast extract, 2.0% peptone, 0.17% yeast nitrogen base with ammonium sulfate, 100 mM potassium phosphate (pH 6.0), 0.4 mg/L biotin, 2.0% MSG, and 2% ethanol. Substitution of ammonium sulphate with 0.5% urea is optional. Carbon source was replenished every 24 h during 72 h induction period. Under these conditions, GFP and RBD yields from PPEPCK equaled and even surpassed those from PAOX1. Compared to the traditional methanol-inducible expression system, the inducers of glutamate-inducible expression system are non-toxic and their metabolism does not generate toxic metabolites such as formaldehyde and hydrogen peroxide. This study sets the stage for MSG-inducible, industrial scale recombinant protein production from K. phaffii PPEPCK in bioreactors.
Keywords: Glutamate inducible expression system; Komagataella phaffii; Monosodium glutamate; PEPCK promoter.
© 2024. The Author(s).
Conflict of interest statement
The intellectual property described in this study is protected by Indian patent No. 432410. U.S. patent is pending (Application No.18/266,357). The glutamate-inducible
Figures
Similar articles
-
The Mut+ strain of Komagataella phaffii (Pichia pastoris) expresses PAOX1 5 and 10 times faster than Muts and Mut- strains: evidence that formaldehyde or/and formate are true inducers of PAOX1.Appl Microbiol Biotechnol. 2020 Sep;104(18):7801-7814. doi: 10.1007/s00253-020-10793-8. Epub 2020 Aug 6. Appl Microbiol Biotechnol. 2020. PMID: 32761464
-
Posttranscriptional regulation of glutamate dehydrogenase 2 and phosphoenolpyruvate carboxykinase in Komagataella phaffii.Yeast. 2022 May;39(5):337-347. doi: 10.1002/yea.3704. Epub 2022 Apr 5. Yeast. 2022. PMID: 35384037
-
Functional recombinant protein is present in the pre-induction phases of Pichia pastoris cultures when grown in bioreactors, but not shake-flasks.Microb Cell Fact. 2014 Sep 4;13(1):127. doi: 10.1186/s12934-014-0127-y. Microb Cell Fact. 2014. PMID: 25186468 Free PMC article.
-
Komagataella phaffii (Pichia pastoris) as a Powerful Yeast Expression System for Biologics Production.Front Biosci (Elite Ed). 2024 Jun 12;16(2):19. doi: 10.31083/j.fbe1602019. Front Biosci (Elite Ed). 2024. PMID: 38939917 Review.
-
Pichia pastoris Promoters.Methods Mol Biol. 2019;1923:97-112. doi: 10.1007/978-1-4939-9024-5_3. Methods Mol Biol. 2019. PMID: 30737736 Review.
References
MeSH terms
Substances
Grants and funding
- JRF/SRF/Human Resource Development Centre, Council of Scientific And Industrial Research
- Fund for Improvement of Science and Technology Infrastructure/Department of Science and Technology, Ministry of Science and Technology, India
- BT/AIR01307/PACE-22-20/Biotechnology Industry Research Assistance Council
- TTR/2022/00023/Science and Engineering Research Board
- CRG/2022/000598/Science and Engineering Research Board
LinkOut - more resources
Full Text Sources
Miscellaneous

