Characterization of the plasma proteome from healthy adult dogs
- PMID: 38638644
- PMCID: PMC11024428
- DOI: 10.3389/fvets.2024.1356318
Characterization of the plasma proteome from healthy adult dogs
Abstract
Introduction: Bloodwork is a widely used diagnostic tool in veterinary medicine, as diagnosis and therapeutic interventions often rely on blood biomarkers. However, biomarkers available in veterinary medicine often lack sensitivity or specificity. Mass spectrometry-based proteomics technology has been extensively used in the analysis of biological fluids. It offers excellent potential for a more comprehensive characterization of the plasma proteome in veterinary medicine.
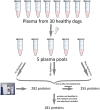
Methods: In this study, we aimed to identify and quantify plasma proteins in a cohort of healthy dogs and compare two techniques for depleting high-abundance plasma proteins to enable the detection of lower-abundance proteins via label-free quantification liquid chromatography-mass spectrometry. We utilized surplus lithium-heparin plasma from 30 healthy dogs, subdivided into five groups of pooled plasma from 6 randomly selected individuals each. Firstly, we used a commercial kit to deplete high-abundance plasma proteins. Secondly, we employed an in-house method to remove albumin using Blue-Sepharose.
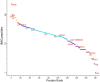
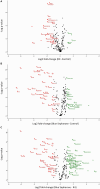
Results and discussion: Among all the samples, some of the most abundant proteins identified were apolipoprotein A and B, albumin, alpha-2-macroglobulin, fibrinogen beta chain, fibronectin, complement C3, serotransferrin, and coagulation factor V. However, neither of the depletion techniques achieved significant depletion of highly abundant proteins. Despite this limitation, we could detect and quantify many clinically relevant proteins. Determining the healthy canine proteome is a crucial first step in establishing a reference proteome for canine plasma. After enrichment, this reference proteome can later be utilized to identify protein markers associated with different diseases, thereby contributing to the diagnosis and prognosis of various pathologies.
Keywords: biomarker; canine; mass-spectrometry; plasma proteomics; veterinary medicine.
Copyright © 2024 Doulidis, Kuropka, Frizzo Ramos, Rodríguez-Rojas and Burgener.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
Similar articles
-
Plasma proteome of buffaloes.Proteomics Clin Appl. 2017 Sep;11(9-10). doi: 10.1002/prca.201600138. Epub 2017 Jun 20. Proteomics Clin Appl. 2017. PMID: 28452126
-
All Cats are Gray in the Dark: Enrichment/Depletion Approaches for Biomarker Discovery on Felis catus Plasma.Proteomics. 2018 Oct;18(20):e1800191. doi: 10.1002/pmic.201800191. Epub 2018 Oct 8. Proteomics. 2018. PMID: 30216667
-
The plasma proteome and the acute phase protein response in canine pyometra.J Proteomics. 2020 Jul 15;223:103817. doi: 10.1016/j.jprot.2020.103817. Epub 2020 May 13. J Proteomics. 2020. PMID: 32416315
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Emerging Affinity-Based Proteomic Technologies for Large-Scale Plasma Profiling in Cardiovascular Disease.Circulation. 2017 Apr 25;135(17):1651-1664. doi: 10.1161/CIRCULATIONAHA.116.025446. Circulation. 2017. PMID: 28438806 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

