Discovering new genes for alfalfa (Medicago sativa) growth and biomass resilience in combined salinity and Phoma medicaginis infection through GWAS
- PMID: 38756967
- PMCID: PMC11096488
- DOI: 10.3389/fpls.2024.1348168
Discovering new genes for alfalfa (Medicago sativa) growth and biomass resilience in combined salinity and Phoma medicaginis infection through GWAS
Abstract
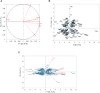
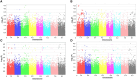
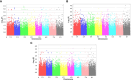
Salinity and Phoma medicaginis infection represent significant challenges for alfalfa cultivation in South Africa, Europe, Australia, and, particularly, Tunisia. These constraints have a severe impact on both yield and quality. The primary aim of this study was to establish the genetic basis of traits associated with biomass and growth of 129 Medicago sativa genotypes through genome-wide association studies (GWAS) under combined salt and P. medicaginis infection stresses. The results of the analysis of variance (ANOVA) indicated that the variation in these traits could be primarily attributed to genotype effects. Among the test genotypes, the length of the main stem, the number of ramifications, the number of chlorotic leaves, and the aerial fresh weight exhibited the most significant variation. The broad-sense heritability (H²) was relatively high for most of the assessed traits, primarily due to genetic factors. Cluster analysis, applied to morpho-physiological traits under the combined stresses, revealed three major groups of accessions. Subsequently, a GWAS analysis was conducted to validate significant associations between 54,866 SNP-filtered single-nucleotide polymorphisms (SNPs) and seven traits. The study identified 27 SNPs that were significantly associated with the following traits: number of healthy leaves (two SNPs), number of chlorotic leaves (five SNPs), number of infected necrotic leaves (three SNPs), aerial fresh weight (six SNPs), aerial dry weight (nine SNPs), number of ramifications (one SNP), and length of the main stem (one SNP). Some of these markers are related to the ionic transporters, cell membrane rigidity (related to salinity tolerance), and the NBS_LRR gene family (associated with disease resistance). These findings underscore the potential for selecting alfalfa genotypes with tolerance to the combined constraints of salinity and P. medicaginis infection.
Keywords: GWAS; Phoma medicaginis; alfalfa; biomass traits; genetic resources; salinity; stress tolerance.
Copyright © 2024 Mnafgui, Jabri, Jihnaoui, Maiza, Guerchi, Zaidi, Basson, Keyster, Djébali, Pecetti, Hanana, Annicchiarico, Sakiroglu, Ludidi and Badri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Genome-Wide Association Study for Major Biofuel Traits in Sorghum Using Minicore Collection.Protein Pept Lett. 2021;28(8):909-928. doi: 10.2174/0929866528666210215141243. Protein Pept Lett. 2021. PMID: 33588716
-
First Report of Spring Black Stem and Leaf Spot of Alfalfa in Washington State Caused by Phoma medicaginis.Plant Dis. 2008 May;92(5):833. doi: 10.1094/PDIS-92-5-0833A. Plant Dis. 2008. PMID: 30769607
-
Arbuscular mycorrhizal fungus alleviates alfalfa leaf spots caused by Phoma medicaginis revealed by RNA-seq analysis.J Appl Microbiol. 2021 Feb;130(2):547-560. doi: 10.1111/jam.14387. Epub 2019 Aug 9. J Appl Microbiol. 2021. PMID: 31310670
-
Alfalfa Spring Black Stem and Leaf Spot Disease Caused by Phoma medicaginis: Epidemic Occurrence and Impacts.Microorganisms. 2024 Jun 24;12(7):1279. doi: 10.3390/microorganisms12071279. Microorganisms. 2024. PMID: 39065048 Free PMC article. Review.
-
Strategies to Increase Prediction Accuracy in Genomic Selection of Complex Traits in Alfalfa (Medicago sativa L.).Cells. 2021 Nov 30;10(12):3372. doi: 10.3390/cells10123372. Cells. 2021. PMID: 34943880 Free PMC article. Review.
References
-
- Ahsan M. Z., Majidano M. S., Bhutto H., Soomro A. W., Panhwar F. H., Channa A. R., et al. . (2015). Genetic variability, coefficient of variance, heritability and genetic advance of some Gossypium hirsutum L. accessions. JAS 7, 147. doi: 10.5539/jas.v7n2p147 - DOI
-
- Annicchiarico P. (2006). Diversity, genetic structure, distinctness and agronomic value of Italian lucerne (Medicago sativa L.) landraces. Euphytica 148, 269–282. doi: 10.1007/s10681-005-9024-0 - DOI
-
- Annicchiarico P., Barrett B., Brummer E. C., Julier B., Marshall A. H. (2015. a). Achievements and challenges in improving temperate perennial forage legumes. Crit. Rev. Plant Sci. 34, 327–380. doi: 10.1080/07352689.2014.898462 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

