Revealing novel and conservative T-cell epitopes with MHC B2 restriction on H9N2 avian influenza virus (AIV)
- PMID: 38768812
- PMCID: PMC11223079
- DOI: 10.1016/j.jbc.2024.107395
Revealing novel and conservative T-cell epitopes with MHC B2 restriction on H9N2 avian influenza virus (AIV)
Abstract
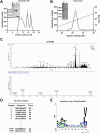
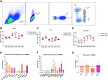
B2 haplotype major histocompatibility complex (MHC) has been extensively reported to confer resistance to various avian diseases. But its peptide-binding motif is unknown, and the presenting peptide is rarely identified. Here, we identified its peptide-binding motif (X-A/V/I/L/P/S/G-X-X-X-X-X-X-V/I/L) in vitro using Random Peptide Library-based MHC I LC-MS/MS analysis. To further clarify the structure basis of motif, we determined the crystal structure of the BF2∗02:01-PB2552-560 complex at 1.9 Å resolution. We found that BF2∗02:01 had a relatively wide antigen-binding groove, and the structural characterization of pockets was consistent with the characterization of peptide-binding motif. The wider features of the peptide-binding motif and increased number of peptides bound by BF2∗02:01 than BF2∗04:01 might resolve the puzzles for the presence of potential H9N2 resistance in B2 chickens. Afterward, we explored the H9N2 avian influenza virus (AIV)-induced cellular immune response in B2 haplotype chickens in vivo. We found that ratio of CD8+ T cell and kinetic expression of cytotoxicity genes including Granzyme K, interferon-γ, NK lysin, and poly-(ADP-ribose) polymerase in peripheral blood mononuclear cells were significantly increased in defending against H9N2 AIV infection. Especially, we selected 425 epitopes as candidate epitopes based on the peptide-binding motif and further identified four CD8+ T-cell epitopes on H9N2 AIV including NS198-106, PB2552-560, NP182-190, and NP455-463 via ELI-spot interferon-γ detections after stimulating memory lymphocytes with peptides. More importantly, these epitopes were found to be conserved in H7N9 AIV and H9N2 AIV. These findings provide direction for developing effective T cell epitope vaccines using well-conserved internal viral antigens in chickens.
Keywords: H9N2 AIV; MHC B2 haplotype; T-cell epitope; crystal structure; peptide-binding motif.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Revealing novel CD8+ T-cell epitopes from the H5N1 avian influenza virus in HBW/B1 haplotype ducks.Vet Res. 2024 Dec 18;55(1):169. doi: 10.1186/s13567-024-01415-6. Vet Res. 2024. PMID: 39695865 Free PMC article.
-
Bursal peptide BP-IV as a novel immunoadjuvant enhances the protective efficacy of an epitope peptide vaccine containing T and B cell epitopes of the H9N2 avian influenza virus.Microb Pathog. 2021 Sep;158:105095. doi: 10.1016/j.micpath.2021.105095. Epub 2021 Jul 16. Microb Pathog. 2021. PMID: 34280501
-
Comparative analysis of key immune protection factors in H9N2 avian influenza viruses infected and immunized specific pathogen-free chicken.Poult Sci. 2021 Jan;100(1):39-46. doi: 10.1016/j.psj.2020.09.080. Epub 2020 Oct 9. Poult Sci. 2021. PMID: 33357705 Free PMC article.
-
Structural Definition of Duck Major Histocompatibility Complex Class I Molecules That Might Explain Efficient Cytotoxic T Lymphocyte Immunity to Influenza A Virus.J Virol. 2017 Jun 26;91(14):e02511-16. doi: 10.1128/JVI.02511-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28490583 Free PMC article.
-
Identification of novel avian influenza virus derived CD8+ T-cell epitopes.PLoS One. 2012;7(2):e31953. doi: 10.1371/journal.pone.0031953. Epub 2012 Feb 23. PLoS One. 2012. PMID: 22384112 Free PMC article.
References
-
- Xu C., Ye H., Qiu W., Lin H., Chen Y., Zhang H., et al. Phylogenetic classification of hemagglutinin gene of H9N2 avian influenza viruses isolated in China during 2012-2016 and evaluation of selected candidate vaccine strains. Poult. Sci. 2018;97:3023–3030. - PubMed
-
- Li Y., Liu M., Sun Q., Zhang H., Zhang H., Jiang S., et al. Genotypic evolution and epidemiological characteristics of H9N2 influenza virus in Shandong Province, China. Poult. Sci. 2019;98:3488–3495. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

