Spiral ganglion neuron degeneration in aminoglycoside-deafened rats involves innate and adaptive immune responses not requiring complement
- PMID: 38840777
- PMCID: PMC11151750
- DOI: 10.3389/fnmol.2024.1389816
Spiral ganglion neuron degeneration in aminoglycoside-deafened rats involves innate and adaptive immune responses not requiring complement
Abstract
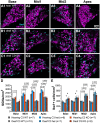
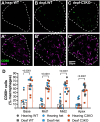
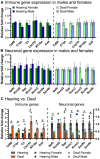
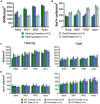
Spiral ganglion neurons (SGNs) transmit auditory information from cochlear hair cells to the brain. SGNs are thus not only important for normal hearing, but also for effective functioning of cochlear implants, which stimulate SGNs when hair cells are missing. SGNs slowly degenerate following aminoglycoside-induced hair cell loss, a process thought to involve an immune response. However, the specific immune response pathways involved remain unknown. We used RNAseq to gain a deeper understanding immune-related and other transcriptomic changes that occur in the rat spiral ganglion after kanamycin-induced deafening. Among the immune and inflammatory genes that were selectively upregulated in deafened spiral ganglia, the complement cascade genes were prominent. We then assessed SGN survival, as well as immune cell numbers and activation, in the spiral ganglia of rats with a CRISPR-Cas9-mediated knockout of complement component 3 (C3). Similar to previous findings in our lab and other deafened rodent models, we observed an increase in macrophage number and increased expression of CD68, a marker of phagocytic activity and cell activation, in macrophages in the deafened ganglia. Moreover, we found an increase in MHCII expression on spiral ganglion macrophages and an increase in lymphocyte number in the deafened ganglia, suggestive of an adaptive immune response. However, C3 knockout did not affect SGN survival or increase in macrophage number/activation, implying that complement activation does not play a role in SGN death after deafening. Together, these data suggest that both innate and adaptive immune responses are activated in the deafened spiral ganglion, with the adaptive response directly contributing to cochlear neurodegeneration.
Keywords: T cell; aminoglycoside ototoxicity; auditory nerve; cochlea; gene expression profiling; inflammation; macrophage; neurodegeneration.
Copyright © 2024 Gansemer, Rahman, Zhang and Green.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
Anti-inflammatory Therapy Protects Spiral Ganglion Neurons After Aminoglycoside Antibiotic-Induced Hair Cell Loss.Neurotherapeutics. 2023 Mar;20(2):578-601. doi: 10.1007/s13311-022-01336-2. Epub 2023 Jan 25. Neurotherapeutics. 2023. PMID: 36697994 Free PMC article.
-
Structural and Ultrastructural Changes to Type I Spiral Ganglion Neurons and Schwann Cells in the Deafened Guinea Pig Cochlea.J Assoc Res Otolaryngol. 2017 Dec;18(6):751-769. doi: 10.1007/s10162-017-0631-y. Epub 2017 Jul 17. J Assoc Res Otolaryngol. 2017. PMID: 28717876 Free PMC article.
-
Postnatal expression of neurotrophic factors accessible to spiral ganglion neurons in the auditory system of adult hearing and deafened rats.J Neurosci. 2014 Sep 24;34(39):13110-26. doi: 10.1523/JNEUROSCI.1014-14.2014. J Neurosci. 2014. PMID: 25253857 Free PMC article.
-
Innate Immunity to Spiral Ganglion Neuron Loss: A Neuroprotective Role of Fractalkine Signaling in Injured Cochlea.Front Cell Neurosci. 2021 Aug 2;15:694292. doi: 10.3389/fncel.2021.694292. eCollection 2021. Front Cell Neurosci. 2021. PMID: 34408629 Free PMC article. Review.
-
Protection of Spiral Ganglion Neurons and Prevention of Auditory Neuropathy.Adv Exp Med Biol. 2019;1130:93-107. doi: 10.1007/978-981-13-6123-4_6. Adv Exp Med Biol. 2019. PMID: 30915703 Review.
References
-
- Alawieh A., Chalhoub R. M., Mallah K., Langley E. F., York M., Broome H., et al. . (2021). Complement drives synaptic degeneration and progressive cognitive decline in the chronic phase after traumatic brain injury. J. Neurosci. 41, 1830–1843. doi: 10.1523/JNEUROSCI.1734-20.2020, PMID: - DOI - PMC - PubMed
-
- Bedeir M. M., Ninoyu Y., Nakamura T., Tsujikawa T., Hirano S. (2022). Multiplex immunohistochemistry reveals cochlear macrophage heterogeneity and local auditory nerve inflammation in cisplatin-induced hearing loss. Front. Neurol. 13:1015014. doi: 10.3389/fneur.2022.1015014, PMID: - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

