Identification of nuclear membrane SUN proteins and components associated with wheat fungal stress responses
- PMID: 38861095
- PMCID: PMC11166608
- DOI: 10.1007/s44154-024-00163-z
Identification of nuclear membrane SUN proteins and components associated with wheat fungal stress responses
Abstract
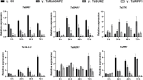
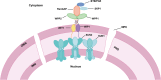
In eukaryotes, the nuclear membrane that encapsulates genomic DNA is composed of an inner nuclear membrane (INM), an outer nuclear membrane (ONM), and a perinuclear space. SUN proteins located in the INM and KASH proteins in the ONM form the SUN-KASH NM-bridge, which functions as the junction of the nucleocytoplasmic complex junction. Proteins containing the SUN domain showed the highest correlation with differentially accumulated proteins (DAPs) in the wheat response to fungal stress. To understand the characteristics of SUN and its associated proteins in wheat responding to pathogen stress, here we investigated and comprehensive analyzed SUN- and KASH-related proteins among the DAPs under fungi infection based on their conserved motifs. In total, four SUN proteins, one WPP domain-interacting protein (WIP), four WPP domain-interacting tail-anchored proteins (WIT), two WPP proteins and one Ran GTPase activating protein (RanGAP) were identified. Following transient expression of Nicotiana benthamiana, TaSUN2, TaRanGAP2, TaWIT1 and TaWIP1 were identified as nuclear membrane proteins, while TaWPP1 and TaWPP2 were expressed in both the nucleus and cell membrane. RT-qPCR analysis demonstrated that the transcription of TaSUN2, TaRanGAP2 and TaWPP1 were strongly upregulated in response to fungal infection. Furthermore, using the bimolecular fluorescence complementation, the luciferase complementation and a nuclear and split-ubiquitin-based membrane yeast two-hybrid systems, we substantiated the interaction between TaSUN2 and TaWIP1, as well as TaWIP1/WIT1 and TaWPP1/WPP2. Silencing of TaSUN2, TaRanGAP2 and TaWPP1 in wheat leaves promoted powdery mildew infection and hyphal growth, and reduced the expression of TaBRI1, TaBAK1 and Ta14-3-3, indicating that these NM proteins play a positive role in resistance to fungal stress. Our study reveals the characteristics of NM proteins and propose the preliminary construction of SUN-WIP-WPP-RanGAP complex in wheat, which represents a foundation for detail elucidating their functions in wheat in future.
Keywords: Triticum aestivum; Fungal stress; KASH; SUN.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
A truncated CC-NB-ARC gene TaRPP13L1-3D positively regulates powdery mildew resistance in wheat via the RanGAP-WPP complex-mediated nucleocytoplasmic shuttle.Planta. 2022 Feb 8;255(3):60. doi: 10.1007/s00425-022-03843-0. Planta. 2022. PMID: 35133503
-
Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination.J Cell Biol. 2012 Jan 23;196(2):203-11. doi: 10.1083/jcb.201108098. J Cell Biol. 2012. PMID: 22270916 Free PMC article.
-
Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1.Nucleus. 2015;6(2):144-53. doi: 10.1080/19491034.2014.1003512. Epub 2015 Mar 11. Nucleus. 2015. PMID: 25759303 Free PMC article.
-
The plant LINC complex at the nuclear envelope.Chromosome Res. 2014 Jun;22(2):241-52. doi: 10.1007/s10577-014-9419-7. Chromosome Res. 2014. PMID: 24801343 Review.
-
_targeting proteins to the plant nuclear envelope.Biochem Soc Trans. 2010 Jun;38(3):733-40. doi: 10.1042/BST0380733. Biochem Soc Trans. 2010. PMID: 20491658 Review.
References
-
- Borgese N, Fasana E (2011) _targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta 1808:937–946. 10.1016/j.bbamem.2010.07.010 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

