This is a preprint.
HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo
- PMID: 38915578
- PMCID: PMC11195054
- DOI: 10.1101/2024.06.10.598329
HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo
Update in
-
HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo.BMC Genomics. 2024 Dec 24;25(1):1240. doi: 10.1186/s12864-024-11162-9. BMC Genomics. 2024. PMID: 39716078 Free PMC article.
Abstract
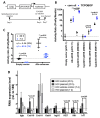
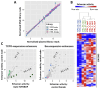
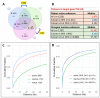
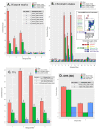
STARR-seq and other massively-parallel reporter assays are widely used to discover functional enhancers in transfected cell models, which can be confounded by plasmid vector-induced type-I interferon immune responses and lack the multicellular environment and endogenous chromatin state of complex mammalian tissues. Here, we describe HDI-STARR-seq, which combines STARR-seq plasmid library delivery to the liver, by hydrodynamic tail vein injection (HDI), with reporter RNA transcriptional initiation driven by a minimal Albumin promoter, which we show is essential for mouse liver STARR-seq enhancer activity assayed 7 days after HDI. Importantly, little or no vector-induced innate type-I interferon responses were observed. Comparisons of HDI-STARR-seq activity between male and female mouse livers and in livers from males treated with an activating ligand of the transcription factor CAR (Nr1i3) identified many condition-dependent enhancers linked to condition-specific gene expression. Further, thousands of active liver enhancers were identified using a high complexity STARR-seq library comprised of ~50,000 genomic regions released by DNase-I digestion of mouse liver nuclei. When compared to stringently inactive library sequences, the active enhancer sequences identified were highly enriched for liver open chromatin regions with activating histone marks (H3K27ac, H3K4me1, H3K4me3), were significantly closer to gene transcriptional start sites, and were significantly depleted of repressive (H3K27me3, H3K9me3) and transcribed region histone marks (H3K36me3). HDI-STARR-seq offers substantial improvements over current methodologies for large scale, functional profiling of enhancers, including condition-dependent enhancers, in liver tissue in vivo, and can be adapted to characterize enhancer activities in a variety of species and tissues by selecting suitable tissue- and species-specific promoter sequences.
Keywords: DNase-seq; MPRA; STARR-seq; TCPOBOP; chromatin accessibility; hydrodynamic tail vein injection; liver epigenetic marks; liver sex differences.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures


Similar articles
-
HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo.Res Sq [Preprint]. 2024 Jun 26:rs.3.rs-4559581. doi: 10.21203/rs.3.rs-4559581/v1. Res Sq. 2024. Update in: BMC Genomics. 2024 Dec 24;25(1):1240. doi: 10.1186/s12864-024-11162-9 PMID: 38978599 Free PMC article. Updated. Preprint.
-
HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo.BMC Genomics. 2024 Dec 24;25(1):1240. doi: 10.1186/s12864-024-11162-9. BMC Genomics. 2024. PMID: 39716078 Free PMC article.
-
Prediction accuracy of regulatory elements from sequence varies by functional sequencing technique.Front Cell Infect Microbiol. 2023 Aug 2;13:1182567. doi: 10.3389/fcimb.2023.1182567. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37600946 Free PMC article.
-
STARR-seq - principles and applications.Genomics. 2015 Sep;106(3):145-150. doi: 10.1016/j.ygeno.2015.06.001. Epub 2015 Jun 11. Genomics. 2015. PMID: 26072434 Review.
-
Principle and application of self-transcribing active regulatory region sequencing in enhancer discovery research.Yi Chuan. 2024 Aug;46(8):589-602. doi: 10.16288/j.yczz.24-149. Yi Chuan. 2024. PMID: 39140141 Review.
References
-
- Shlyueva D, Stampfel G, Stark A: Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014, 15(4):272–286. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
