Feeder-free culture of human pluripotent stem cells drives MDM4-mediated gain of chromosome 1q
- PMID: 38964325
- PMCID: PMC11368687
- DOI: 10.1016/j.stemcr.2024.06.003
Feeder-free culture of human pluripotent stem cells drives MDM4-mediated gain of chromosome 1q
Abstract
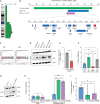
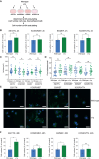
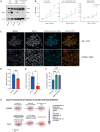
Culture-acquired variants in human pluripotent stem cells (hPSCs) hinder their applications in research and clinic. However, the mechanisms that underpin selection of variants remain unclear. Here, through analysis of comprehensive karyotyping datasets from over 23,000 hPSC cultures of more than 1,500 lines, we explored how culture conditions shape variant selection. Strikingly, we identified an association of chromosome 1q gains with feeder-free cultures and noted a rise in its prevalence in recent years, coinciding with increased usage of feeder-free regimens. Competition experiments of multiple isogenic lines with and without a chromosome 1q gain confirmed that 1q variants have an advantage in feeder-free (E8/vitronectin), but not feeder-based, culture. Mechanistically, we show that overexpression of MDM4, located on chromosome 1q, drives variants' advantage in E8/vitronectin by alleviating genome damage-induced apoptosis, which is lower in feeder-based conditions. Our study explains condition-dependent patterns of hPSC aberrations and offers insights into the mechanisms of variant selection.
Keywords: MDM4; culture conditions; genetic changes; genome damage; human pluripotent stem cells.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.E.L. is a co-inventor and receives a share of royalties on various hPSC media- and culture-related patents currently owned and licensed by the Wisconsin Alumni Research Foundation (WARF). I.B. is a member of the scientific advisory board of WiCell. U.B.-D. received consulting fees from Accent Therapeutics.
Figures

Similar articles
-
Gain of 1q confers an MDM4-driven growth advantage to undifferentiated and differentiating hESC while altering their differentiation capacity.Cell Death Dis. 2024 Nov 21;15(11):852. doi: 10.1038/s41419-024-07236-x. Cell Death Dis. 2024. PMID: 39572522 Free PMC article.
-
MDM4 Is _targeted by 1q Gain and Drives Disease in Burkitt Lymphoma.Cancer Res. 2019 Jun 15;79(12):3125-3138. doi: 10.1158/0008-5472.CAN-18-3438. Epub 2019 Apr 18. Cancer Res. 2019. PMID: 31000522
-
Robust protocol for feeder-free adaptation of cryopreserved human pluripotent stem cells.In Vitro Cell Dev Biol Anim. 2019 Dec;55(10):777-783. doi: 10.1007/s11626-019-00413-9. Epub 2019 Oct 29. In Vitro Cell Dev Biol Anim. 2019. PMID: 31664691
-
Concise review: The evolution of human pluripotent stem cell culture: from feeder cells to synthetic coatings.Stem Cells. 2013 Jan;31(1):1-7. doi: 10.1002/stem.1260. Stem Cells. 2013. PMID: 23081828 Free PMC article. Review.
-
Development of substrates for the culture of human pluripotent stem cells.Biomater Sci. 2023 May 2;11(9):2974-2987. doi: 10.1039/d2bm01473d. Biomater Sci. 2023. PMID: 37009904 Review.
Cited by
-
Gain of 1q confers an MDM4-driven growth advantage to undifferentiated and differentiating hESC while altering their differentiation capacity.Cell Death Dis. 2024 Nov 21;15(11):852. doi: 10.1038/s41419-024-07236-x. Cell Death Dis. 2024. PMID: 39572522 Free PMC article.
-
De Novo Cancer Mutations Frequently Associate with Recurrent Chromosomal Abnormalities during Long-Term Human Pluripotent Stem Cell Culture.Cells. 2024 Aug 21;13(16):1395. doi: 10.3390/cells13161395. Cells. 2024. PMID: 39195283 Free PMC article.
References
-
- Andrews P.W., Barbaric I., Benvenisty N., Draper J.S., Ludwig T., Merkle F.T., Sato Y., Spits C., Stacey G.N., Wang H., Pera M.F. The consequences of recurrent genetic and epigenetic variants in human pluripotent stem cells. Cell Stem Cell. 2022;29:1624–1636. doi: 10.1016/j.stem.2022.11.006. - DOI - PubMed
-
- Andrews P.W., Ben-David U., Benvenisty N., Coffey P., Eggan K., Knowles B.B., Nagy A., Pera M., Reubinoff B., Rugg-Gunn P.J., Stacey G.N. Assessing the Safety of Human Pluripotent Stem Cells and Their Derivatives for Clinical Applications. Stem Cell Rep. 2017;9:1–4. doi: 10.1016/j.stemcr.2017.05.029. - DOI - PMC - PubMed
-
- Avery S., Hirst A.J., Baker D., Lim C.Y., Alagaratnam S., Skotheim R.I., Lothe R.A., Pera M.F., Colman A., Robson P., et al. BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Rep. 2013;1:379–386. doi: 10.1016/j.stemcr.2013.10.005. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

