P300 regulates histone crotonylation and preimplantation embryo development
- PMID: 39080296
- PMCID: PMC11289097
- DOI: 10.1038/s41467-024-50731-0
P300 regulates histone crotonylation and preimplantation embryo development
Abstract
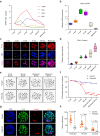
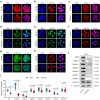
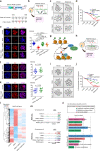
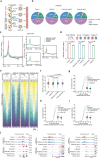
Histone lysine crotonylation, an evolutionarily conserved modification differing from acetylation, exerts pivotal control over diverse biological processes. Among these are gene transcriptional regulation, spermatogenesis, and cell cycle processes. However, the dynamic changes and functions of histone crotonylation in preimplantation embryonic development in mammals remain unclear. Here, we show that the transcription coactivator P300 functions as a writer of histone crotonylation during embryonic development. Depletion of P300 results in significant developmental defects and dysregulation of the transcriptome of embryos. Importantly, we demonstrate that P300 catalyzes the crotonylation of histone, directly stimulating transcription and regulating gene expression, thereby ensuring successful progression of embryo development up to the blastocyst stage. Moreover, the modification of histone H3 lysine 18 crotonylation (H3K18cr) is primarily localized to active promoter regions. This modification serves as a distinctive epigenetic indicator of crucial transcriptional regulators, facilitating the activation of gene transcription. Together, our results propose a model wherein P300-mediated histone crotonylation plays a crucial role in regulating the fate of embryonic development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Quantitative Crotonylome Analysis Expands the Roles of p300 in the Regulation of Lysine Crotonylation Pathway.Proteomics. 2018 Aug;18(15):e1700230. doi: 10.1002/pmic.201700230. Epub 2018 Jul 11. Proteomics. 2018. PMID: 29932303 Free PMC article.
-
Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation.Mol Cell. 2015 Apr 16;58(2):203-15. doi: 10.1016/j.molcel.2015.02.029. Epub 2015 Mar 26. Mol Cell. 2015. PMID: 25818647 Free PMC article.
-
A Chemical Probe for Protein Crotonylation.J Am Chem Soc. 2018 Apr 11;140(14):4757-4760. doi: 10.1021/jacs.7b13141. Epub 2018 Apr 2. J Am Chem Soc. 2018. PMID: 29584949 Free PMC article.
-
Histone crotonylation-centric gene regulation.Epigenetics Chromatin. 2021 Feb 6;14(1):10. doi: 10.1186/s13072-021-00385-9. Epigenetics Chromatin. 2021. PMID: 33549150 Free PMC article. Review.
-
Lysine crotonylation: A challenging new player in the epigenetic regulation of plants.J Proteomics. 2022 Mar 20;255:104488. doi: 10.1016/j.jprot.2022.104488. Epub 2022 Jan 20. J Proteomics. 2022. PMID: 35065287 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

