Bioinformatics for Inosine: Tools and Approaches to Trace This Elusive RNA Modification
- PMID: 39202357
- PMCID: PMC11353476
- DOI: 10.3390/genes15080996
Bioinformatics for Inosine: Tools and Approaches to Trace This Elusive RNA Modification
Abstract
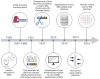
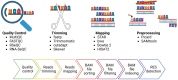
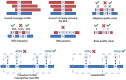
Inosine is a nucleotide resulting from the deamination of adenosine in RNA. This chemical modification process, known as RNA editing, is typically mediated by a family of double-stranded RNA binding proteins named Adenosine Deaminase Acting on dsRNA (ADAR). While the presence of ADAR orthologs has been traced throughout the evolution of metazoans, the existence and extension of RNA editing have been characterized in a more limited number of animals so far. Undoubtedly, ADAR-mediated RNA editing plays a vital role in physiology, organismal development and disease, making the understanding of the evolutionary conservation of this phenomenon pivotal to a deep characterization of relevant biological processes. However, the lack of direct high-throughput methods to reveal RNA modifications at single nucleotide resolution limited an extended investigation of RNA editing. Nowadays, these methods have been developed, and appropriate bioinformatic pipelines are required to fully exploit this data, which can complement existing approaches to detect ADAR editing. Here, we review the current literature on the "bioinformatics for inosine" subject and we discuss future research avenues in the field.
Keywords: A-to-I editing; ADAR; RNA editing; bioinformatics; inosine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Adenosine-to-Inosine RNA Editing Enzyme ADAR and microRNAs.Methods Mol Biol. 2021;2181:83-95. doi: 10.1007/978-1-0716-0787-9_6. Methods Mol Biol. 2021. PMID: 32729076
-
The role of dsRNA A-to-I editing catalyzed by ADAR family enzymes in the pathogeneses.RNA Biol. 2024 Jan;21(1):52-69. doi: 10.1080/15476286.2024.2414156. Epub 2024 Oct 24. RNA Biol. 2024. PMID: 39449182 Free PMC article. Review.
-
A-to-I RNA editing and hematopoiesis.Exp Hematol. 2024 Nov;139:104621. doi: 10.1016/j.exphem.2024.104621. Epub 2024 Aug 24. Exp Hematol. 2024. PMID: 39187172 Review.
-
Adenosine deaminases that act on RNA, then and now.RNA. 2024 Apr 16;30(5):521-529. doi: 10.1261/rna.079990.124. RNA. 2024. PMID: 38531651 Free PMC article.
-
An I for an A: Dynamic Regulation of Adenosine Deamination-Mediated RNA Editing.Genes (Basel). 2021 Jul 1;12(7):1026. doi: 10.3390/genes12071026. Genes (Basel). 2021. PMID: 34356042 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

