Expanding the inhibitor space of the WWP1 and WWP2 HECT E3 ligases
- PMID: 39223706
- PMCID: PMC11373361
- DOI: 10.1080/14756366.2024.2394895
Expanding the inhibitor space of the WWP1 and WWP2 HECT E3 ligases
Abstract
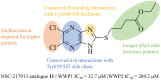
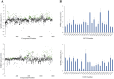
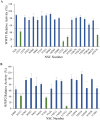
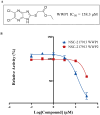
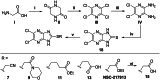
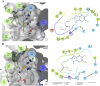
The HECT E3 ubiquitin ligases 1 (WWP1) and 2 (WWP2) are responsible for the ubiquitin-mediated degradation of key tumour suppressor proteins and are dysregulated in various cancers and diseases. Here we expand their limited inhibitor space by identification of NSC-217913 displaying a WWP1 IC50 of 158.3 µM (95% CI = 128.7, 195.1 µM). A structure-activity relationship by synthesis approach aided by molecular docking led to compound 11 which displayed increased potency with an IC50 of 32.7 µM (95% CI = 24.6, 44.3 µM) for WWP1 and 269.2 µM (95% CI = 209.4, 347.9 µM) for WWP2. Molecular docking yielded active site-bound poses suggesting that the heterocyclic imidazo[4,5-b]pyrazine scaffold undertakes a π-stacking interaction with the phenolic group of tyrosine, and the ethyl ester enables strong ion-dipole interactions. Given the therapeutic potential of WWP1 and WWP2, we propose that compound 11 may provide a basis for future lead compound development.
Keywords: SAR; WWP1; WWP2; drug discovery; ubiquitin ligase inhibitors.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
E2UbcH5B-derived peptide ligands _target HECT E3-E2 binding site and block the Ub-dependent SARS-CoV-2 egression: A computational study.Comput Biol Med. 2022 Jul;146:105660. doi: 10.1016/j.compbiomed.2022.105660. Epub 2022 May 22. Comput Biol Med. 2022. PMID: 35751189 Free PMC article.
-
Discovery of Small Molecule WWP2 Ubiquitin Ligase Inhibitors.Chemistry. 2018 Dec 3;24(67):17677-17680. doi: 10.1002/chem.201804169. Epub 2018 Nov 6. Chemistry. 2018. PMID: 30207403
-
A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases.Nat Commun. 2019 Jul 18;10(1):3162. doi: 10.1038/s41467-019-11224-7. Nat Commun. 2019. PMID: 31320636 Free PMC article.
-
The role of WWP1 and WWP2 in bone/cartilage development and diseases.Mol Cell Biochem. 2024 Nov;479(11):2907-2919. doi: 10.1007/s11010-023-04917-7. Epub 2024 Jan 22. Mol Cell Biochem. 2024. PMID: 38252355 Review.
-
WWP1 E3 ligase at the crossroads of health and disease.Cell Death Dis. 2023 Dec 21;14(12):853. doi: 10.1038/s41419-023-06380-0. Cell Death Dis. 2023. PMID: 38129384 Free PMC article. Review.
References
-
- Komander D, Rape M.. The ubiquitin code. Annu Rev Biochem. 2012;81(1):203–229. - PubMed
-
- Kane RC, Bross PF, Farrell AT, Pazdur R.. Velcade®: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8(6):508–513. - PubMed
-
- Kwon YT, Ciechanover A.. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. 2017;42(11):873–886. - PubMed
-
- Sharma A, Preuss CV. Bortezomib [Internet]. Treasure Island (FL): StatPearls Publishing; 2024. [cited 2024 Mar 21]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK519559.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
