Viral gene drive spread during herpes simplex virus 1 infection in mice
- PMID: 39289368
- PMCID: PMC11408514
- DOI: 10.1038/s41467-024-52395-2
Viral gene drive spread during herpes simplex virus 1 infection in mice
Abstract
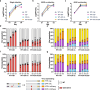
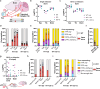
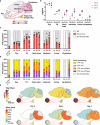
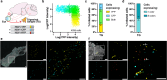
Gene drives are genetic modifications designed to propagate efficiently through a population. Most applications rely on homologous recombination during sexual reproduction in diploid organisms such as insects, but we recently developed a gene drive in herpesviruses that relies on co-infection of cells by wild-type and engineered viruses. Here, we report on a viral gene drive against human herpes simplex virus 1 (HSV-1) and show that it propagates efficiently in cell culture and during HSV-1 infection in mice. We describe high levels of co-infection and gene drive-mediated recombination in neuronal tissues during herpes encephalitis as the infection progresses from the site of inoculation to the peripheral and central nervous systems. In addition, we show evidence that a superinfecting gene drive virus could recombine with wild-type viruses during latent infection. These findings indicate that HSV-1 achieves high rates of co-infection and recombination during viral infection, a phenomenon that is currently underappreciated. Overall, this study shows that a viral gene drive could spread in vivo during HSV-1 infection, paving the way toward therapeutic applications.
© 2024. The Author(s).
Conflict of interest statement
A patent application describing the use of a gene drive in DNA viruses has been filed by the Buck Institute for Research on Aging (Application number 17054760, inventor: M.W.). The authors declare no further competing interests.
Figures




Similar articles
-
Herpes Simplex Virus 1 Coinfection Modifies Adeno-associated Virus Genome End Recombination.J Virol. 2021 Jun 10;95(13):e0048621. doi: 10.1128/JVI.00486-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853961 Free PMC article.
-
A dual fluorescent herpes simplex virus type 1 recombinant reveals divergent outcomes of neuronal infection.J Virol. 2024 May 14;98(5):e0003224. doi: 10.1128/jvi.00032-24. Epub 2024 Apr 23. J Virol. 2024. PMID: 38651900 Free PMC article.
-
Herpes Simplex Virus 1 Replication, Ocular Disease, and Reactivations from Latency Are Restricted Unilaterally after Inoculation of Virus into the Lip.J Virol. 2019 Nov 26;93(24):e01586-19. doi: 10.1128/JVI.01586-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554680 Free PMC article.
-
Roles of Us8A and Its Phosphorylation Mediated by Us3 in Herpes Simplex Virus 1 Pathogenesis.J Virol. 2016 May 27;90(12):5622-5635. doi: 10.1128/JVI.00446-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27030266 Free PMC article.
-
Herpes simplex virus latent infection in the nervous system.J Neurovirol. 1995 Mar;1(1):19-29. doi: 10.3109/13550289509111007. J Neurovirol. 1995. PMID: 9222339 Review.
References
-
- Brown, E. L. et al. Effect of maternal herpes simplex virus (HSV) serostatus and HSV type on risk of neonatal herpes. Acta Obstet. Gynecol. Scand.86, 523–529 (2007). - PubMed
-
- Freeman, E. E. et al. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS20, 73–83 (2006). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

