Potent neutralization of SARS-CoV-2 variants by RBD nanoparticle and prefusion-stabilized spike immunogens
- PMID: 39379400
- PMCID: PMC11461925
- DOI: 10.1038/s41541-024-00982-1
Potent neutralization of SARS-CoV-2 variants by RBD nanoparticle and prefusion-stabilized spike immunogens
Abstract
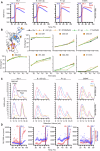
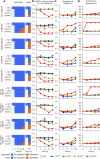
We previously described a two-component protein nanoparticle vaccine platform that displays 60 copies of the SARS-CoV-2 spike protein RBD (RBD-NP). The vaccine, when adjuvanted with AS03, was shown to elicit robust neutralizing antibody and CD4 T cell responses in Phase I/II clinical trials, met its primary co-endpoints in a Phase III trial, and has been licensed by multiple regulatory authorities under the brand name SKYCovioneTM. Here we characterize the biophysical properties, stability, antigenicity, and immunogenicity of RBD-NP immunogens incorporating mutations from the B.1.351 (β) and P.1 (γ) variants of concern (VOCs) that emerged in 2020. We also show that the RBD-NP platform can be adapted to the Omicron strains BA.5 and XBB.1.5. We compare β and γ variant and E484K point mutant nanoparticle immunogens to the nanoparticle displaying the Wu-1 RBD, as well as to soluble prefusion-stabilized (HexaPro) spike trimers harboring VOC-derived mutations. We find the properties of immunogens based on different SARS-CoV-2 variants can differ substantially, which could affect the viability of variant vaccine development. Introducing stabilizing mutations in the linoleic acid binding site of the RBD-NPs resulted in increased physical stability compared to versions lacking the stabilizing mutations without deleteriously affecting immunogenicity. The RBD-NP immunogens and HexaPro trimers, as well as combinations of VOC-based immunogens, elicited comparable levels of neutralizing antibodies against distinct VOCs. Our results demonstrate that RBD-NP-based vaccines can elicit neutralizing antibody responses against SARS-CoV-2 variants and can be rapidly designed and stabilized, demonstrating the potential of two-component RBD-NPs as a platform for the development of broadly protective coronavirus vaccines.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
A.C.W., D.E., D.V., and N.P.K. are named as inventors on patent applications filed by the University of Washington based on the studies presented in this paper. The King lab has received unrelated sponsored research agreements from Pfizer and GSK. D.V. is a consultant for and has received an unrelated sponsored research agreement from Vir Biotechnology Inc. P.K. and A.P. were employees and shareholders of Kymab Ltd, now Kymab, a Sanofi Company. The other authors declare no competing interests.
Figures



Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
SARS-CoV-2 BA.4/5 infection triggers more cross-reactive FcγRIIIa signaling and neutralization than BA.1, in the context of hybrid immunity.J Virol. 2024 Jul 23;98(7):e0067824. doi: 10.1128/jvi.00678-24. Epub 2024 Jul 2. J Virol. 2024. PMID: 38953380 Free PMC article.
-
Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines.bioRxiv [Preprint]. 2021 Mar 16:2021.03.15.435528. doi: 10.1101/2021.03.15.435528. bioRxiv. 2021. Update in: Cell. 2021 Oct 14;184(21):5432-5447.e16. doi: 10.1016/j.cell.2021.09.015 PMID: 33758839 Free PMC article. Updated. Preprint.
-
Safety and immunogenicity of a modified mRNA-lipid nanoparticle vaccine candidate against COVID-19: Results from a phase 1, dose-escalation study.Hum Vaccin Immunother. 2024 Dec 31;20(1):2408863. doi: 10.1080/21645515.2024.2408863. Epub 2024 Oct 18. Hum Vaccin Immunother. 2024. PMID: 39422261 Free PMC article. Clinical Trial.
-
Strategies to improve smoking cessation rates in primary care.Cochrane Database Syst Rev. 2021 Sep 6;9(9):CD011556. doi: 10.1002/14651858.CD011556.pub2. Cochrane Database Syst Rev. 2021. PMID: 34693994 Free PMC article. Review.
References
-
- Novel Coronavirus – China. https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON233.
-
- Tegally, H. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature592, 438–443 (2021). - PubMed
Grants and funding
- R01 GM120553/GM/NIGMS NIH HHS/United States
- DP1 AI158186/AI/NIAID NIH HHS/United States
- P01 AI167966/AI/NIAID NIH HHS/United States
- HHSN272201700059C/AI/NIAID NIH HHS/United States
- P01AI167966/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

