This is a preprint.
Cell-type-specific mapping of enhancers and _target genes from single-cell multimodal data
- PMID: 39386519
- PMCID: PMC11463474
- DOI: 10.1101/2024.09.24.614814
Cell-type-specific mapping of enhancers and _target genes from single-cell multimodal data
Abstract
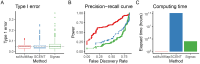
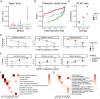
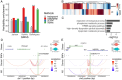
Mapping enhancers and _target genes in disease-related cell types has provided critical insights into the functional mechanisms of genetic variants identified by genome-wide association studies (GWAS). However, most existing analyses rely on bulk data or cultured cell lines, which may fail to identify cell-type-specific enhancers and _target genes. Recently, single-cell multimodal data measuring both gene expression and chromatin accessibility within the same cells have enabled the inference of enhancer-gene pairs in a cell-type-specific and context-specific manner. However, this task is challenged by the data's high sparsity, sequencing depth variation, and the computational burden of analyzing a large number of enhancer-gene pairs. To address these challenges, we propose scMultiMap, a statistical method that infers enhancer-gene association from sparse multimodal counts using a joint latent-variable model. It adjusts for technical confounding, permits fast moment-based estimation and provides analytically derived -values. In systematic analyses of blood and brain data, scMultiMap shows appropriate type I error control, high statistical power with greater reproducibility across independent datasets and stronger consistency with orthogonal data modalities. Meanwhile, its computational cost is less than 1% of existing methods. When applied to single-cell multimodal data from postmortem brain samples from Alzheimer's disease (AD) patients and controls, scMultiMap gave the highest heritability enrichment in microglia and revealed new insights into the regulatory mechanisms of AD GWAS variants in microglia.
Keywords: cell-type-specific analysis; enhancer; latent-variable model; single-cell multimodal data.
Figures

Similar articles
-
Mapping enhancer and chromatin accessibility landscapes charts the regulatory network of Alzheimer's disease.Comput Biol Med. 2024 Jan;168:107802. doi: 10.1016/j.compbiomed.2023.107802. Epub 2023 Dec 3. Comput Biol Med. 2024. PMID: 38056211
-
Enhancer variants associated with Alzheimer's disease affect gene expression via chromatin looping.BMC Med Genomics. 2019 Sep 9;12(1):128. doi: 10.1186/s12920-019-0574-8. BMC Med Genomics. 2019. PMID: 31500627 Free PMC article.
-
Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants.Proc Natl Acad Sci U S A. 2013 Oct 29;110(44):17921-6. doi: 10.1073/pnas.1317023110. Epub 2013 Oct 14. Proc Natl Acad Sci U S A. 2013. PMID: 24127591 Free PMC article.
-
Genetic insights into immune mechanisms of Alzheimer's and Parkinson's disease.Front Immunol. 2023 Jun 8;14:1168539. doi: 10.3389/fimmu.2023.1168539. eCollection 2023. Front Immunol. 2023. PMID: 37359515 Free PMC article. Review.
-
Enhancer Dysfunction in 3D Genome and Disease.Cells. 2019 Oct 19;8(10):1281. doi: 10.3390/cells8101281. Cells. 2019. PMID: 31635067 Free PMC article. Review.
References
-
- Mostafavi H., Spence J. P., Naqvi S. & Pritchard J. K. Systematic differences in discovery of genetic effects on gene expression and complex traits. Nature Genetics 55, 1866–1875 (2023). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
