Parvimonas micra forms a distinct bacterial network with oral pathobionts in colorectal cancer patients
- PMID: 39420333
- PMCID: PMC11487773
- DOI: 10.1186/s12967-024-05720-8
Parvimonas micra forms a distinct bacterial network with oral pathobionts in colorectal cancer patients
Abstract
Background: Mounting evidence suggests a significant role of the gut microbiota in the development and progression of colorectal cancer (CRC). In particular, an over-representation of oral pathogens has been linked to CRC. The aim of this study was to further investigate the faecal microbial landscape of CRC patients, with a focus on the oral pathogens Parvimonas micra and Fusobacterium nucleatum.
Methods: In this study, 16S rRNA sequencing was conducted using faecal samples from CRC patients (n = 275) and controls without pathological findings (n = 95).
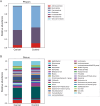
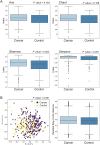
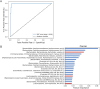
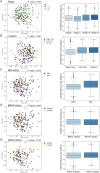
Results: We discovered a significant difference in microbial composition depending on tumour location and microsatellite instability (MSI) status, with P. micra, F. nucleatum, and Peptostreptococcus stomatis found to be more abundant in patients with MSI tumours. Moreover, P. micra and F. nucleatum were associated with a cluster of CRC-related bacteria including Bacteroides fragilis as well as with other oral pathogens such as P. stomatis and various Porphyromonas species. This cluster was distinctly different in the control group, suggesting its potential linkage with CRC.
Conclusions: Our results suggest a similar distribution of several CRC-associated bacteria within CRC patients, underscoring the importance of considering the concomitant presence of bacterial species in studies investigating the mechanisms of CRC development and progression.
Keywords: Fusobacterium nucelatum; Parvimonas micra; Colorectal cancer; Intestinal microbiota; Oral pathobionts.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer.Sci Rep. 2021 Feb 3;11(1):2925. doi: 10.1038/s41598-021-82465-0. Sci Rep. 2021. PMID: 33536501 Free PMC article.
-
Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer.Sci Rep. 2020 Sep 17;10(1):15250. doi: 10.1038/s41598-020-72132-1. Sci Rep. 2020. PMID: 32943695 Free PMC article.
-
Parvimonas micra can translocate from the subgingival sulcus of the human oral cavity to colorectal adenocarcinoma.Mol Oncol. 2024 May;18(5):1143-1173. doi: 10.1002/1878-0261.13506. Epub 2023 Sep 13. Mol Oncol. 2024. PMID: 37558206 Free PMC article.
-
The role of anaerobic bacteria in the development and prevention of colorectal cancer: A review study.Anaerobe. 2022 Feb;73:102501. doi: 10.1016/j.anaerobe.2021.102501. Epub 2021 Dec 12. Anaerobe. 2022. PMID: 34906686 Review.
-
Colorectal cancer and gut microbiota studies in China.Gut Microbes. 2023 Jan-Dec;15(1):2236364. doi: 10.1080/19490976.2023.2236364. Gut Microbes. 2023. PMID: 37482657 Free PMC article. Review.
References
-
- Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. - PubMed
-
- Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–82. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

