Therapeutic peptides identification via kernel risk sensitive loss-based k-nearest neighbor model and multi-Laplacian regularization
- PMID: 39438076
- PMCID: PMC11495874
- DOI: 10.1093/bib/bbae534
Therapeutic peptides identification via kernel risk sensitive loss-based k-nearest neighbor model and multi-Laplacian regularization
Abstract
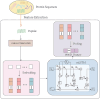
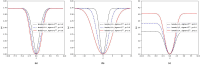
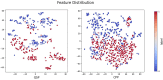
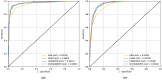
Therapeutic peptides are therapeutic agents synthesized from natural amino acids, which can be used as carriers for precisely transporting drugs and can activate the immune system for preventing and treating various diseases. However, screening therapeutic peptides using biochemical assays is expensive, time-consuming, and limited by experimental conditions and biological samples, and there may be ethical considerations in the clinical stage. In contrast, screening therapeutic peptides using machine learning and computational methods is efficient, automated, and can accurately predict potential therapeutic peptides. In this study, a k-nearest neighbor model based on multi-Laplacian and kernel risk sensitive loss was proposed, which introduces a kernel risk loss function derived from the K-local hyperplane distance nearest neighbor model as well as combining the Laplacian regularization method to predict therapeutic peptides. The findings indicated that the suggested approach achieved satisfactory results and could effectively predict therapeutic peptide sequences.
Keywords: Laplacian regularized model; kernel risk-sensitive mean p-power error; multi-information fusion; sequence classification; therapeutic peptides.
© The Author(s) 2024. Published by Oxford University Press.
Figures



Similar articles
-
Identify ncRNA Subcellular Localization via Graph Regularized k-Local Hyperplane Distance Nearest Neighbor Model on Multi-Kernel Learning.IEEE/ACM Trans Comput Biol Bioinform. 2022 Nov-Dec;19(6):3517-3529. doi: 10.1109/TCBB.2021.3107621. Epub 2022 Dec 8. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 34432632
-
Multi-view local hyperplane nearest neighbor model based on independence criterion for identifying vesicular transport proteins.Int J Biol Macromol. 2023 Aug 30;247:125774. doi: 10.1016/j.ijbiomac.2023.125774. Epub 2023 Jul 13. Int J Biol Macromol. 2023. PMID: 37437677
-
UMPred-FRL: A New Approach for Accurate Prediction of Umami Peptides Using Feature Representation Learning.Int J Mol Sci. 2021 Dec 4;22(23):13124. doi: 10.3390/ijms222313124. Int J Mol Sci. 2021. PMID: 34884927 Free PMC article.
-
Combining non-negative matrix factorization with graph Laplacian regularization for predicting drug-miRNA associations based on multi-source information fusion.Front Pharmacol. 2023 Feb 2;14:1132012. doi: 10.3389/fphar.2023.1132012. eCollection 2023. Front Pharmacol. 2023. PMID: 36817132 Free PMC article.
-
Laplacian embedded regression for scalable manifold regularization.IEEE Trans Neural Netw Learn Syst. 2012 Jun;23(6):902-15. doi: 10.1109/TNNLS.2012.2190420. IEEE Trans Neural Netw Learn Syst. 2012. PMID: 24806762
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

