Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase
- PMID: 9223306
- PMCID: PMC21548
- DOI: 10.1073/pnas.94.15.8016
Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase
Abstract
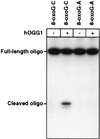
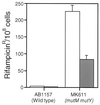
The major mutagenic base lesion in DNA caused by exposure to reactive oxygen species is 8-hydroxyguanine (8-oxo-7, 8-dihydroguanine). In bacteria and Saccharomyces cerevisiae, this damaged base is excised by a DNA glycosylase with an associated lyase activity for chain cleavage. We have cloned, sequenced, and expressed a human cDNA with partial sequence homology to the relevant yeast gene. The encoded 47-kDa human enzyme releases free 8-hydroxyguanine from oxidized DNA and introduces a chain break in a double-stranded oligonucleotide specifically at an 8-hydroxyguanine residue base paired with cytosine. Expression of the human protein in a DNA repair-deficient E. coli mutM mutY strain partly suppresses its spontaneous mutator phenotype. The gene encoding the human enzyme maps to chromosome 3p25. These results show that human cells have an enzyme that can initiate base excision repair at mutagenic DNA lesions caused by active oxygen.
Figures



Similar articles
-
Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 1997 Jul 22;94(15):8010-5. doi: 10.1073/pnas.94.15.8010. Proc Natl Acad Sci U S A. 1997. PMID: 9223305 Free PMC article.
-
Excision repair of 8-hydroxyguanine in mammalian cells: the mouse Ogg1 protein as a model.Free Radic Res. 1998 Dec;29(6):487-97. doi: 10.1080/10715769800300541. Free Radic Res. 1998. PMID: 10098454
-
Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine.Proc Natl Acad Sci U S A. 1996 May 28;93(11):5197-202. doi: 10.1073/pnas.93.11.5197. Proc Natl Acad Sci U S A. 1996. PMID: 8643552 Free PMC article.
-
The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis.Antioxid Redox Signal. 2001 Aug;3(4):597-609. doi: 10.1089/15230860152542952. Antioxid Redox Signal. 2001. PMID: 11554447 Review.
-
The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis.Arch Biochem Biophys. 2000 May 1;377(1):1-8. doi: 10.1006/abbi.2000.1773. Arch Biochem Biophys. 2000. PMID: 10775435 Review.
Cited by
-
Deficiency of base excision repair enzyme NEIL3 drives increased predisposition to autoimmunity.J Clin Invest. 2016 Nov 1;126(11):4219-4236. doi: 10.1172/JCI85647. Epub 2016 Oct 17. J Clin Invest. 2016. PMID: 27760045 Free PMC article.
-
Dynamic relocalization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant.Nucleic Acids Res. 2005 Mar 30;33(6):1813-24. doi: 10.1093/nar/gki325. Print 2005. Nucleic Acids Res. 2005. PMID: 15800211 Free PMC article.
-
Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo.Mol Cell Biol. 2006 Mar;26(5):1654-65. doi: 10.1128/MCB.26.5.1654-1665.2006. Mol Cell Biol. 2006. PMID: 16478987 Free PMC article.
-
The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage.Mol Cell Biol. 2004 Jul;24(13):6084-93. doi: 10.1128/MCB.24.13.6084-6093.2004. Mol Cell Biol. 2004. PMID: 15199162 Free PMC article.
-
Base excision repair of reactive oxygen species-initiated 7,8-dihydro-8-oxo-2'-deoxyguanosine inhibits the cytotoxicity of platinum anticancer drugs.Mol Cancer Ther. 2009 Jul;8(7):2015-26. doi: 10.1158/1535-7163.MCT-08-0929. Epub 2009 Jun 30. Mol Cancer Ther. 2009. PMID: 19567822 Free PMC article.
References
-
- Lindahl T. Nature (London) 1993;362:709–715. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

