Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association
- PMID: 9442102
- PMCID: PMC2132567
- DOI: 10.1083/jcb.140.2.259
Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association
Abstract
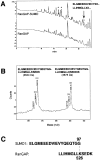
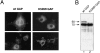
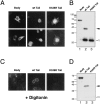
The mammalian guanosine triphosphate (GTP)ase-activating protein RanGAP1 is the first example of a protein covalently linked to the ubiquitin-related protein SUMO-1. Here we used peptide mapping, mass spectroscopy analysis, and mutagenesis to identify the nature of the link between RanGAP1 and SUMO-1. SUMO-1 is linked to RanGAP1 via glycine 97, indicating that the last 4 amino acids of this 101- amino acid protein are proteolytically removed before its attachment to RanGAP1. Recombinant SUMO-1 lacking the last four amino acids is efficiently used for modification of RanGAP1 in vitro and of multiple unknown proteins in vivo. In contrast to most ubiquitinated proteins, only a single lysine residue (K526) in RanGAP1 can serve as the acceptor site for modification by SUMO-1. Modification of RanGAP1 with SUMO-1 leads to association of RanGAP1 with the nuclear envelope (NE), where it was previously shown to be required for nuclear protein import. Sufficient information for modification and _targeting resides in a 25-kD domain of RanGAP1. RanGAP1-SUMO-1 remains stably associated with the NE during many cycles of in vitro import. This indicates that removal of RanGAP1 from the NE is not a required element of nuclear protein import and suggests that the reversible modification of RanGAP1 may have a regulatory role.
Figures




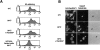

Similar articles
-
SUMO-1 modification and its role in _targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex.J Cell Biol. 1998 Feb 9;140(3):499-509. doi: 10.1083/jcb.140.3.499. J Cell Biol. 1998. PMID: 9456312 Free PMC article.
-
A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex.J Cell Biol. 1996 Dec;135(6 Pt 1):1457-70. doi: 10.1083/jcb.135.6.1457. J Cell Biol. 1996. PMID: 8978815 Free PMC article.
-
A small ubiquitin-related polypeptide involved in _targeting RanGAP1 to nuclear pore complex protein RanBP2.Cell. 1997 Jan 10;88(1):97-107. doi: 10.1016/s0092-8674(00)81862-0. Cell. 1997. PMID: 9019411
-
SUMO/sentrin: protein modifiers regulating important cellular functions.Biochem Cell Biol. 1999;77(4):299-309. Biochem Cell Biol. 1999. PMID: 10546893 Review.
-
SUMO, ubiquitin's mysterious cousin.Nat Rev Mol Cell Biol. 2001 Mar;2(3):202-10. doi: 10.1038/35056591. Nat Rev Mol Cell Biol. 2001. PMID: 11265250 Review.
Cited by
-
Sea urchin vault structure, composition, and differential localization during development.BMC Dev Biol. 2005 Feb 14;5:3. doi: 10.1186/1471-213X-5-3. BMC Dev Biol. 2005. PMID: 15710043 Free PMC article.
-
An in vitro Förster resonance energy transfer-based high-throughput screening assay identifies inhibitors of SUMOylation E2 Ubc9.Acta Pharmacol Sin. 2020 Nov;41(11):1497-1506. doi: 10.1038/s41401-020-0405-7. Epub 2020 Apr 27. Acta Pharmacol Sin. 2020. PMID: 32341466 Free PMC article.
-
Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains.Mol Cell Biol. 2002 Aug;22(16):5708-20. doi: 10.1128/MCB.22.16.5708-5720.2002. Mol Cell Biol. 2002. PMID: 12138183 Free PMC article.
-
Emerging roles of the SUMO pathway in development.Cell Mol Life Sci. 2011 Dec;68(24):4045-64. doi: 10.1007/s00018-011-0792-5. Epub 2011 Sep 4. Cell Mol Life Sci. 2011. PMID: 21892772 Free PMC article. Review.
-
MicroRNA-mediated regulation of Ubc9 expression in cancer cells.Clin Cancer Res. 2009 Mar 1;15(5):1550-7. doi: 10.1158/1078-0432.CCR-08-0820. Epub 2009 Feb 17. Clin Cancer Res. 2009. PMID: 19223510 Free PMC article.
References
-
- Adam SA, Sterne-Marr R, Gerace L. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 1992;219:97–110. - PubMed
-
- Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271:2823–2831. - PubMed
-
- Baldi L, Brown K, Franzoso G, Siebenlist U. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of I κ B-α. J Biol Chem. 1996;271:376–379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

