Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2
- PMID: 9621066
- PMCID: PMC110408
- DOI: 10.1128/JVI.72.7.6034-6039.1998
Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2
Abstract
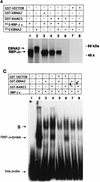
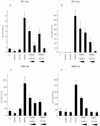
The intracellular region (RAMIC) of the mouse Notch1 receptor interacts with RBP-J/CBF-1, which binds to the DNA sequence CGTGGGAA and suppresses differentiation by transcriptional activation of genes regulated by RBP-J. Epstein-Barr virus nuclear antigen 2 (EBNA2) is essential for immortalization of human B cells by the virus. EBNA2 is a pleiotropic activator of viral and cellular genes and is _targeted to DNA at least in part by interacting with RBP-J. We found that EBNA2 and the Notch1 RAMIC compete for binding to RBP-J, indicating that their interaction sites on RBP-J overlap at least partially. EBNA2 and Notch1 RAMIC transactivated the same set of viral and host promoters, i.e., the EBNA2 response element of the Epstein-Barr virus TP1 and the HES-1 promoter. Furthermore, EBNA2 functionally replaced the Notch1 RAMIC by suppressing differentiation of C2C12 myoblast progenitor cells.
Figures



Similar articles
-
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa.Immunobiology. 1997 Dec;198(1-3):299-306. doi: 10.1016/s0171-2985(97)80050-2. Immunobiology. 1997. PMID: 9442401 Review.
-
Intracellular forms of human NOTCH1 interact at distinctly different levels with RBP-jkappa in human B and T cells.Leukemia. 2000 Jan;14(1):84-92. doi: 10.1038/sj.leu.2401630. Leukemia. 2000. PMID: 10637481
-
Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters.J Virol. 1999 Apr;73(4):2770-80. doi: 10.1128/JVI.73.4.2770-2780.1999. J Virol. 1999. PMID: 10074124 Free PMC article.
-
Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2.J Virol. 2000 Feb;74(4):1727-35. doi: 10.1128/jvi.74.4.1727-1735.2000. J Virol. 2000. PMID: 10644343 Free PMC article.
-
EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes.Semin Cancer Biol. 2001 Dec;11(6):423-34. doi: 10.1006/scbi.2001.0409. Semin Cancer Biol. 2001. PMID: 11669604 Review.
Cited by
-
The expression and function of Epstein-Barr virus encoded latent genes.Mol Pathol. 2000 Oct;53(5):238-47. doi: 10.1136/mp.53.5.238. Mol Pathol. 2000. PMID: 11091847 Free PMC article. Review.
-
A role for SKIP in EBNA2 activation of CBF1-repressed promoters.J Virol. 2000 Feb;74(4):1939-47. doi: 10.1128/jvi.74.4.1939-1947.2000. J Virol. 2000. PMID: 10644367 Free PMC article.
-
Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus.J Virol. 2001 Jul;75(13):5899-912. doi: 10.1128/JVI.75.13.5899-5912.2001. J Virol. 2001. PMID: 11390591 Free PMC article.
-
EBF1 binds to EBNA2 and promotes the assembly of EBNA2 chromatin complexes in B cells.PLoS Pathog. 2017 Oct 2;13(10):e1006664. doi: 10.1371/journal.ppat.1006664. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28968461 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway.J Virol. 2005 Mar;79(6):3468-78. doi: 10.1128/JVI.79.6.3468-3478.2005. J Virol. 2005. PMID: 15731241 Free PMC article.
References
-
- Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. - PubMed
-
- Aster J C, Robertson E S, Hasserjian R B, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. - PubMed
-
- Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

