Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor
- PMID: 9864345
- PMCID: PMC103564
- DOI: 10.1128/JB.181.1.319-330.1999
Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor
Abstract
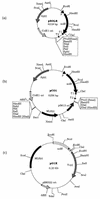
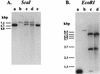
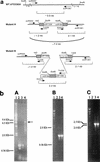
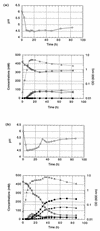
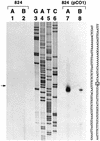
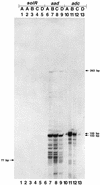
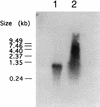
A gene (orf1, now designated solR) previously identified upstream of the aldehyde/alcohol dehydrogenase gene aad (R. V. Nair, G. N. Bennett, and E. T. Papoutsakis, J. Bacteriol. 176:871-885, 1994) was found to encode a repressor of the sol locus (aad, ctfA, ctfB and adc) genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824. Primer extension analysis identified a transcriptional start site 35 bp upstream of the solR start codon. Amino acid comparisons of SolR identified a potential helix-turn-helix DNA-binding motif in the C-terminal half towards the center of the protein, suggesting a regulatory role. Overexpression of SolR in strain ATCC 824(pCO1) resulted in a solvent-negative phenotype owing to its deleterious effect on the transcription of the sol locus genes. Inactivation of solR in C. acetobutylicum via homologous recombination yielded mutants B and H (ATCC 824 solR::pO1X) which exhibited deregulated solvent production characterized by increased flux towards butanol and acetone formation, earlier induction of aad, lower overall acid production, markedly improved yields of solvents on glucose, a prolonged solvent production phase, and increased biomass accumulation compared to those of the wild-type strain.
Figures


Similar articles
-
Small and Low but Potent: the Complex Regulatory Role of the Small RNA SolB in Solventogenesis in Clostridium acetobutylicum.Appl Environ Microbiol. 2018 Jul 2;84(14):e00597-18. doi: 10.1128/AEM.00597-18. Print 2018 Jul 15. Appl Environ Microbiol. 2018. PMID: 29728392 Free PMC article.
-
Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene.J Ind Microbiol Biotechnol. 2001 Nov;27(5):322-8. doi: 10.1038/sj.jim.7000191. J Ind Microbiol Biotechnol. 2001. PMID: 11781808
-
Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824.J Bacteriol. 1994 Feb;176(3):871-85. doi: 10.1128/jb.176.3.871-885.1994. J Bacteriol. 1994. PMID: 8300540 Free PMC article.
-
Transcriptional regulation of solventogenesis in Clostridium acetobutylicum.J Mol Microbiol Biotechnol. 2002 May;4(3):295-300. J Mol Microbiol Biotechnol. 2002. PMID: 11931561 Review.
-
Novel and neglected issues of acetone-butanol-ethanol (ABE) fermentation by clostridia: Clostridium metabolic diversity, tools for process mapping and continuous fermentation systems.Biotechnol Adv. 2013 Jan-Feb;31(1):58-67. doi: 10.1016/j.biotechadv.2012.01.010. Epub 2012 Jan 28. Biotechnol Adv. 2013. PMID: 22306328 Review.
Cited by
-
Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824.J Bacteriol. 2002 Jul;184(13):3586-97. doi: 10.1128/JB.184.13.3586-3597.2002. J Bacteriol. 2002. PMID: 12057953 Free PMC article.
-
Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824.Appl Environ Microbiol. 2003 May;69(5):2831-41. doi: 10.1128/AEM.69.5.2831-2841.2003. Appl Environ Microbiol. 2003. PMID: 12732555 Free PMC article.
-
Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces.Appl Environ Microbiol. 2000 Mar;66(3):1107-13. doi: 10.1128/AEM.66.3.1107-1113.2000. Appl Environ Microbiol. 2000. PMID: 10698778 Free PMC article.
-
The path to next generation biofuels: successes and challenges in the era of synthetic biology.Microb Cell Fact. 2010 Jan 20;9:3. doi: 10.1186/1475-2859-9-3. Microb Cell Fact. 2010. PMID: 20089184 Free PMC article. Review.
-
Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways.Nat Chem Biol. 2011 Apr;7(4):222-7. doi: 10.1038/nchembio.537. Epub 2011 Feb 27. Nat Chem Biol. 2011. PMID: 21358636
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

