Two polymorphic variants of wild-type p53 differ biochemically and biologically
- PMID: 9891044
- PMCID: PMC116039
- DOI: 10.1128/MCB.19.2.1092
Two polymorphic variants of wild-type p53 differ biochemically and biologically
Abstract
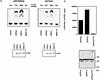
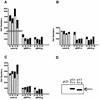
The wild-type p53 protein exhibits a common polymorphism at amino acid 72, resulting in either a proline residue (p53Pro) or an arginine residue (p53Arg) at this position. Despite the difference that this change makes in the primary structure of the protein resulting in a difference in migration during sodium dodecyl sulfate-polyacrylamide gel electrophoresis, no differences in the biochemical or biological characteristics of these wild-type p53 variants have been reported. We have recently shown that p53Arg is significantly more susceptible than p53Pro to the degradation induced by human papillomavirus (HPV) E6 protein. Moreover, this may result in an increased susceptibility to HPV-induced tumors in homozygous p53Arg individuals. In further investigating the characteristics of these p53 variants, we now show that both forms are morphologically wild type and do not differ in their ability to bind to DNA in a sequence-specific manner. However, there are a number of differences between the p53 variants in their abilities to bind components of the transcriptional machinery, to activate transcription, to induce apoptosis, and to repress the transformation of primary cells. These observations may have implications for the development of cancers which harbor wild-type p53 sequences and possibly for the ability of such tumors to respond to therapy, depending on their p53 genotype.
Figures






Similar articles
-
p53 codon 72 polymorphism and risk of cervical carcinoma in Korean women.J Korean Med Sci. 2000 Feb;15(1):65-7. doi: 10.3346/jkms.2000.15.1.65. J Korean Med Sci. 2000. PMID: 10719811 Free PMC article.
-
Human papillomavirus type 16 E6 variants in cervical carcinoma: relationship to host genetic factors and clinical parameters.J Gen Virol. 1999 Dec;80 ( Pt 12):3233-3240. doi: 10.1099/0022-1317-80-12-3233. J Gen Virol. 1999. PMID: 10567656
-
Mutant p53 can substitute for human papillomavirus type 16 E6 in immortalization of human keratinocytes but does not have E6-associated trans-activation or transforming activity.J Virol. 1992 Jul;66(7):4201-8. doi: 10.1128/JVI.66.7.4201-4208.1992. J Virol. 1992. PMID: 1318401 Free PMC article.
-
The role of the E6-p53 interaction in the molecular pathogenesis of HPV.Oncogene. 1999 Dec 13;18(53):7690-700. doi: 10.1038/sj.onc.1202953. Oncogene. 1999. PMID: 10618709 Review.
-
The p53 tumor suppressor gene and gene product.Princess Takamatsu Symp. 1989;20:221-30. Princess Takamatsu Symp. 1989. PMID: 2488233 Review.
Cited by
-
Impact of codon 72 Arg > Pro single nucleotide polymorphism in TP53 gene in the risk of kangri cancer: a case control study in Kashmir.Tumour Biol. 2012 Aug;33(4):927-33. doi: 10.1007/s13277-012-0318-2. Epub 2012 Jan 17. Tumour Biol. 2012. PMID: 22249977
-
Expression of pAkt affects p53 codon 72 polymorphism-based prediction of response to radiotherapy in nasopharyngeal carcinoma.Radiat Oncol. 2013 May 11;8:117. doi: 10.1186/1748-717X-8-117. Radiat Oncol. 2013. PMID: 23663243 Free PMC article.
-
TP53 codon 72 Gene Polymorphism Paradox in Associated with Various Carcinoma Incidences, Invasiveness and Chemotherapy Responses.Int J Biomed Sci. 2008 Dec;4(4):248-54. Int J Biomed Sci. 2008. PMID: 23675098 Free PMC article.
-
Unique profile of ordered arrangements of repetitive elements in the C57BL/6J mouse genome implicating their functional roles.PLoS One. 2012;7(4):e35156. doi: 10.1371/journal.pone.0035156. Epub 2012 Apr 18. PLoS One. 2012. PMID: 22529984 Free PMC article.
-
p53 codon 72 polymorphism association with head and neck squamous cell carcinoma.Braz J Otorhinolaryngol. 2010 May-Jun;76(3):316-20. doi: 10.1590/S1808-86942010000300008. Braz J Otorhinolaryngol. 2010. PMID: 20658010 Free PMC article.
References
-
- Banks L, Matlashewski G, Crawford L. Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986;159:529–534. - PubMed
-
- Beckman G, Birgander R, Sjalander A, Saha N, Holmberg P, Kiveld S, Beckman L. Is p53 polymorphism maintained by natural selection. Hum Hered. 1994;44:266–270. - PubMed
-
- Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-_target genes. Nature. 1994;370:220–223. - PubMed
-
- Cho Y, Gorina S, Jeffrey P, Pavletich N. Crystal structure of a p53 tumour suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. - PubMed
-
- Donehower L, Harvey B, Slagle B, McArthur B, Montgomery C, Butel J, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
