Abstract
Objective
To propose an algorithm that utilizes only timestamps of longitudinal electronic health record data to classify clinical deterioration events.
Materials and methods
This retrospective study explores the efficacy of machine learning algorithms in classifying clinical deterioration events among patients in intensive care units using sequences of timestamps of vital sign measurements, flowsheets comments, order entries, and nursing notes. We design a data pipeline to partition events into discrete, regular time bins that we refer to as timesteps. Logistic regressions, random forest classifiers, and recurrent neural networks are trained on datasets of different length of timesteps, respectively, against a composite outcome of death, cardiac arrest, and Rapid Response Team calls. Then these models are validated on a holdout dataset.
Results
A total of 6720 intensive care unit encounters meet the criteria and the final dataset includes 830 578 timestamps. The gated recurrent unit model utilizes timestamps of vital signs, order entries, flowsheet comments, and nursing notes to achieve the best performance on the time-to-outcome dataset, with an area under the precision-recall curve of 0.101 (0.06, 0.137), a sensitivity of 0.443, and a positive predictive value of 0. 092 at the threshold of 0.6.
Discussion and Conclusion
This study demonstrates that our recurrent neural network models using only timestamps of longitudinal electronic health record data that reflect healthcare processes achieve well-performing discriminative power.
Keywords: electronic health records; predictive modeling; clinical informatics; early warning scores, machine learning
INTRODUCTION
Delayed recognition and failure to provide timely intervention to patients developing clinical deterioration events result in suboptimal prognoses.1,2 Patients often develop physiological instability hours before clinical deterioration,3–5 and thus many are potentially avoidable.6 Early warning score systems have been developed to assist clinicians in recognizing early signs of physiological deterioration, allowing them to provide timely intervention.7 Early warning scores (EWS) typically take physiological measurements to calculate risk scores of clinical deteriorations.8–12 Different levels of action are recommended to corresponding healthcare professionals if a patient exceeds certain thresholds.13 These actions typically include increasing monitoring and examinations, initiating treatments, and eventually activating the Rapid Response Team to elevate patient care. A Rapid Response Team (RRT) is a group of clinicians that respond to patients with early signs of deterioration to prevent respiratory and cardiac arrest in the hospital.14,15 The implementations of EWS and RRT are associated with decreased mortality and in-hospital cardiac arrest.16–19 In recent reviews of EWS, most data-driven EWS use structured data (eg, vital signs, lab data, demography) from electronic health records (EHR),20,21 but a few include clinical concern where clinicians answer structured questions if he/she is subjectively concerned about the patient.22,23 In clinical practice, experienced clinicians recognize patterns of patient deterioration based on expert knowledge and clinical assessment skills and from “knowing the patient” rather than solely on objective physiological measurements.24–26 These subjective expert judgements, often made before any physiological derangement is detected by machines, are captured in EHR data.25,27–29
It is widely known that the EHR is not a direct reflection of patient’s pathophysiology but rather a record of the healthcare process with noise.30,31 Time is an important component that represents the dynamic and positive feedback loop of healthcare processes.30,31 Additional measurements and clinical records are often made when clinicians are concerned about their patients’ conditions.25,27 For example, a series of vital sign measures in early morning implies a different healthcare process than a set of those recorded as a routine measurement at scheduled times. Therefore, features reflecting the healthcare process (eg, time and co-occurrence of measurements, order placement, prescriptions, notes) can likewise be utilized for model building. In the National Institute for Nursing Research-funded Communicating Narrative Concerns Entered by RNs (CONCERN) study, our team demonstrates that the inclusion of documentation and measurement frequency in an EWS triggered an alert 5–26 hours earlier than the first Modified Early Warning Score (MEWS) alert.32 In our more recent work, the CONCERNv2.0 model,, including additional nursing note features and temporal features, further increases the “lead time” to 42 hours compared to MEWS and National Early Warning Score.33 Most EWSs rely solely on physiological measurements, but we believe the use of features that represent clinicians’ concerns could augment early detection.
We propose an algorithm that utilizes only timestamps of longitudinal EHR data (eg, time and co-occurrence of vital sign measurements, flowsheet comments, order entries, and nursing notes) to classify clinical deterioration events. These time-series data reflect nurses’ decision-making related to patient surveillance.25,28 We emphasize that our data for analysis do not include any measurement values (eg, heart rate = 90mHg). This study aims to 1) validate the proposed models built on sequences of timestamps of underlying clinical data that reflect the healthcare process, and 2) evaluate the impact of including time-of-day and time-to-outcome information in the model.
MATERIALS AND METHODS
This project was approved by both Columbia University Irving Medical Center and Brigham and Women's Hospital Institutional Review Board for the Protection of Human Subjects.
This retrospective study explores the efficacy of machine learning algorithms in classifying clinical deterioration among patients in intensive care units (ICU) using sequences of timestamps of vital sign measurements, flowsheets comments, order entries, and nursing notes in electronic health records. We design a data pipeline to partition events into discrete, regular time bins that we refer to as timesteps. Logistic regressions, random forest classifiers, and recurrent neural network (RNN) algorithms are tested on a training dataset against a composite outcome of death, cardiac arrest, and RRT calls. We choose single layer Long Short-Term Memory (LSTM) and Gated Recurrent Unit (GRU), 2 well-established modalities, as our RNN classifiers.34,35 Then we validate our algorithms on a holdout dataset with Area Under the Precision-Recall Curve (AUPRC) sensitivity, Positive Predictive Value (PPV), and F-score. The code is accessible on the online repository: https://github.com/lf2608/Timestamps_EHR_Deterioration_Predict.
Cohort selection
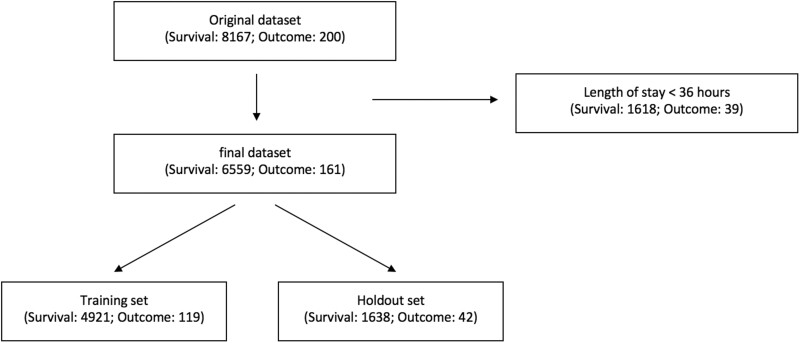
This study utilizes the deidentified dataset of ICU admissions from a composite site of ICUs within a healthcare system in the northeast region of US (Supplementary Material, Appendix 1). We select a subset of patients admitted during a 4-year period from 2016 to January 2019. Each site has its own go-live date to collect data, based on the dates that the commercial EHR system Epic went live in 2016. Inclusion criteria for the patient cohort are: 1) at least 18 years of age at the time of admission and 2) non-hospice and non-palliative care patients.9 We exclude hospice and palliative care patients since these admissions have different care plans and healthcare processes compared to non-palliative care. Clinicians focus more on patient comfort rather than disease progression; therefore, fewer clinical measurements and documentations are made for hospice and palliative care patients. We select the first ICU visit (via admission or transfer) for a given patient’s hospital encounter. ICU stays shorter than 36 hours are excluded (Figure 1) because we require at least 24 hours of data for analysis followed by a 12-hour time span to predict an outcome. As part of the deidentification process, the date of each ICU stay is shifted randomly to a range between Apr 2013 to Feb 2017 but in a way that the month, day of week and time stay the same. We randomly split the dataset into a training set (75%) and a holdout set (25%). The holdout set is only used for model validation.
Figure 1.
Cohort selection. There are 8367 unique ICU admissions including 200 outcome events in this dataset. After removing ICU admissions shorter than 36 hours, the final dataset consists of 6720 ICU admissions. (Table 3) There are 161 admissions with composite outcomes of death, cardiac arrest, or Rapid Response Team call, which consist of 2.40% of the dataset. Finally, we split the dataset into training set and holdout set in a ratio of 3:1.
Feature selection
We select timestamps from 15 types of underlying clinical data, namely vital signs, flowsheets comments, order entry, and nursing notes for analysis. These chosen clinical data have the following characteristics: 1) regularly collected and entered into EHR on all patients, 2) reflecting clinicians’ judgement or decision-making process based on patients’ conditions.25,27,28 These timestamps represent the time when the data was collected by the clinician, but not the time when it was entered into the EHR. The method of feature selection is reported in our previous works.32,33 (Table 1) The partitioning of data which results in the creation of timesteps will be further explained in the Data Preprocessing section.
Table 1.
Categories of features used in model derivation
| Category | Features |
|---|---|
| Vital signs entered* | heart rate (HR), blood pressure (BP), respiratory rate (RR), body temperature (BT), oxygen saturation (SpO2) |
| Flowsheet comments | heart rate comments, blood pressure comments, respiratory rate comments, body temperature comments, oxygen saturation comments |
| Order entries | single vital sign measurement, complete set of vital signs measurement, one-time medication order, medication withholds |
| Nursing notes | documentation made by nurses |
Any automated device that generates data requires a nurse’s manual validation during data collection.
Primary outcome
The primary outcome of this study is a composite outcome of death, cardiac arrest, and RRT calls. The policy for initiating RRT calls at our study sites is as follows: nurses make decisions to activate RRT based on established early warning criteria related to the patient’s respiratory, cardiovascular, neurologic, and other conditions. Death, cardiac arrest, and RRT calls are the measurable clinical events that occur for patients who experience deterioration. We choose this composite outcome in order to increase the prevalence of the outcome set. If more than 1 outcome occurred in an ICU stay, the first event is defined as time of primary outcome.
Data preprocessing pipeline
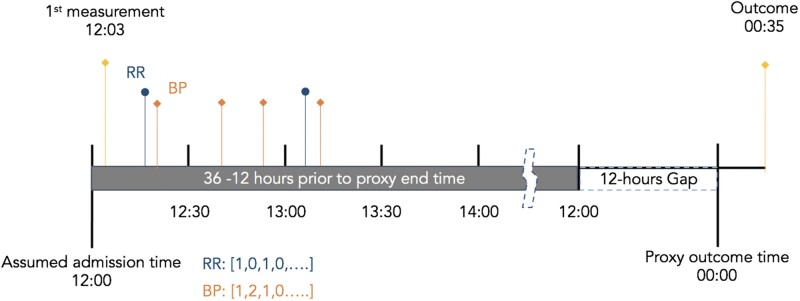
The exact time of ICU admission is not accessible from the database. With the assumption that the admission time is approximated by the first record of data, our data preprocessing pipeline rounds down the time of first record to the nearest hour and uses that as the admission time. Likewise, it rounds down the outcome time to the nearest hour for ease of computation. It then extracts data from 12 to 36 hours prior to discharge (n = 6559) or the following composite outcome of death, cardiac arrest, or RRT call (n = 161) for final analysis. Next, it partitions the 24-hour sequence into regularly spaced timesteps and calculates the count of records for every feature within each timestep (Figure 2). Each timestep is indexed by the corresponding time-of-day. Then the pipeline sorts the data by the time-of-day index so the first row of the data always starts at 12 am (Table 2). The indexes are then dropped from the dataset so they are not used for model derivation. In our dataset, discharges happened mostly during the day while deterioration events occurred more evenly between day and night (Supplementary Material Appendix 6). The time-of-day index of data’s first timestep is decided by the hour of outcome since we only sample data 36 to 12 hours prior to that. Rather than creating an additional variable which leaks the information about the time of outcome, this method embeds the time-of-day information by sorting the data by the time-of-day index. Our pipeline also generates the unsorted dataset by keeping the time-to-outcome sequence of the healthcare process. In the following text, we call the sorted dataset the “time-of-day dataset” and the unsorted the “time-to-outcome dataset.”
Figure 2.
Time series partitioned by a given unit of time in data preprocessing. Our data pipeline rounds down both the admission time and outcome time to the nearest earlier hour for the ease of computation. It then partitions the sequences of timestamps into regularly spaced timesteps and calculates the count of records for every feature within each timestep. Data 36 to 12 hours prior to the outcomes are used for final analysis.
Table 2.
Feature representation of the time-of-day dataset
| Time-of-day Index | Timesteps Index |
Feature A | Feature B | Feature C | Feature D | ….. | Feature K |
|---|---|---|---|---|---|---|---|
| [00:00, 00:30) | 25 | 1 | 0 | 0 | 0 | …… | 0 |
| …… | …… | …… | …… | …… | …… | …… | …… |
| [12:00, 12:30) | 1 | 0 | 3 | 1 | 0 | ….. | 2 |
| [12:30, 13:00) | 2 | 5 | 4 | 1 | 3 | …… | 1 |
| [13:00, 13:30) | 3 | 0 | 0 | 0 | 0 | …… | 0 |
| [13:30, 14:00) | 4 | 1 | 0 | 1 | 0 | …… | 1 |
| …… | ….. | …… | …… | …… | …… | …… | …… |
| [23:30, 00:00) | 24 | 0 | 0 | 1 | 1 | …… | 0 |
This table illustrates the feature representation of an ICU stay. In this example, the original data starts at time-of-day 12 pm and is then sorted by the time-of-day index. The interpretation for the first row, the 25th timestep of the data: there is 1 measurement of Feature A during the time-of-day [00:00, 00:30]. The indexes are not used for model derivation.
We visualize the distribution of timestamps of vital signs, flowsheet comments, order entries, and nursing notes in the dataset against time-of-day. While notes have a bimodal distribution, major clusters on the hour and smaller clusters on the quarter of hour are shown for vital signs, data entries, and flowsheet comments. Most interevent intervals of the features are within 2 hours long (Supplementary Material Appendix 4). Therefore, we set 3 different lengths of timesteps (15, 30, 60 minutes) for partition in an attempt to capture different levels of granularity in the timestamp sequences. For algorithms taking a single data point (ie, logistic regression and random forest classifier), we calculate the count of timestamps for each feature over the 24 hours analytic period.
Model derivation and validation
We apply the oversampling technique on the training set to tackle the imbalanced data during model training. In order to optimize discriminating power as well as to prevent overfitting, we set a dictionary of hyperparameters for each classifier in our model selection pipeline. The dictionary includes L1 and L2 regularization for logistic regression,36,37 pruning parameters for random forest classifier, as well as parameters for number of neurons, learning rate, and dropout layer before outputs of LSTM and GRU algorithm (Supplementary Material Appendix 2). The pipeline returns the best logistic regression and random forest classifier with optimal hyperparameters using grid search approach and 4-fold cross-validation from python package scikit-learn (v.0.23.2). It also optimizes the hyperparameters for the single layer LSTM and GRU algorithm (tensorflow v.2.3.1) using the hyperband technique from python package keras-tuner (v.1.0.1). Hyperband is a bandit-based approach to speed up random search through adaptive resource allocation and early stopping.38 We then set the thresholds for these classifiers with the aim of achieving a well-performing sensitivity while retaining an acceptable PPV. In the literature, PPV is often _targeted in the range of 10%–20% in model validation in order to avoid excessive false alarms.39–41 The LSTM and GRU model are trained on a time-series dataset of different length of timesteps (15, 30, 60 minutes) while logistic regression and random forest classifiers are trained on overall counts of timestamps. Finally, we validate these classifiers on a holdout set based on AUPRC, sensitivity, PPV, and F-score since the ICU stays with outcomes are rare. We then utilize bootstrapping to estimate the 95% confidence interval of the model.
RESULTS
There are 8367 unique ICU admissions including 200 outcome events in this dataset. These are broken into 76 cardiac arrest and 124 RRT calls. Since these were the first outcome events that the patients had, patients who developed cardiac arrest then died were recorded as cardiac arrest in this dataset. After removing ICU admissions shorter than 36 hours, the final dataset consists of 6720 ICU admissions. (Table 3) There are 161 samples with composite outcomes of death, cardiac arrest, or RRT call which consist of 2.40% of the dataset. A total of 830 578 timestamps are included in the final analysis (Supplementary Material Appendix 3).
Table 3.
Characteristics of study complete dataset
| Admissions | Counts/Values | |||
|---|---|---|---|---|
| Unique ICU admission, n | 6720 | |||
| Age at admission, years-old |
Range: 19–89 Mean: 64.8 Quartiles Values (57, 67, 76) |
|||
| Male, n (%) | 3931 (58.5%) | |||
| First adverse event in episode | ||||
| None, n (%) | 6559 (97.60%) | |||
| Composite outcomes, n (%) | 161 (2.40%) | |||
|
| ||||
| Observations | Total Counts | Unique Patient Counts | Average Counts per Patient | STD |
|
| ||||
| Heart rate entered | 175 812 | 6446 | 26.16 | 10.45 |
| Respiratory rate entered | 137 342 | 6302 | 20.44 | 11.27 |
| Blood pressure entered | 156 471 | 6441 | 23.28 | 10.32 |
| Body temperature entered | 65 705 | 6448 | 9.78 | 8.71 |
| Oxygen saturation entered | 159 431 | 6443 | 23.72 | 11.53 |
| Single vital sign measurement | 10 851 | 3502 | 1.61 | 4.16 |
| Complete set of vital signs measurement | 47 210 | 6174 | 7.03 | 7.12 |
| One-time medication order | 34 653 | 6078 | 5.16 | 4.26 |
| Medication withhold | 26 154 | 5886 | 3.89 | 3.33 |
| Heart rate comments | 308 | 237 | 0.05 | 0.28 |
| Respiratory rate comments | 120 | 103 | 0.02 | 0.16 |
| Blood pressure comments | 1140 | 708 | 0.17 | 0.70 |
| Body temperature comments | 136 | 104 | 0.02 | 0.21 |
| Oxygen saturation comments | 351 | 296 | 0.05 | 0.27 |
| Notes | 14 894 | 5972 | 2.22 | 1.49 |
The GRU model, utilizing timestamps of vital signs, order entries, flowsheet comments, and nursing notes on the time-to-outcome dataset of timesteps of 60 minutes, has the best predictive power with an AUPRC of 0.101 (0.06, 0.137). This model performs better than the logistic regression with L2 normalization AUPRC 0.093 (0.09, 0.096), but the difference is not statistically significant. (Table 4) The best GRU model has a sensitivity of 0.443 and a PPV of 0.092 at the threshold of 0.6. In general, the RNN models on time-to-outcome datasets have better AUPRC than those on time-of-day datasets. The performance of the RNN models varies on datasets of different length of timesteps with no significant differences.
Table 4.
Model validation
| Algorithm | Length of Timestep | AUPRC (95 CI) |
F-Score (95 CI) |
Sensitivity (95 CI) |
PPV (95 CI) |
Specificity (95 CI) |
NPV (95 CI) |
AUROC (95 CI) |
|---|---|---|---|---|---|---|---|---|
| Time-of-day Dataset | ||||||||
| GRU | 60 | 0.082 (0.051, 0.108) | 0.122 (0.05, 0.156) | 0.354 (0.035, 0.524) | 0.084 (0.053, 0.15) | 0.888 (0.794, 0.992) | 0.982 (0.976, 0.985) | 0.7 (0.656, 0.724) |
| GRU | 30 | 0.074 (0.046, 0.102) | 0.118 (0.0, 0.157) | 0.376 (0.0, 0.548) | 0.076 (0.0, 0.122) | 0.88 (0.751, 0.996) | 0.982 (0.975, 0.986) | 0.703 (0.627, 0.733) |
| GRU | 15 | 0.07 (0.044, 0.097) | 0.118 (0.025, 0.152) | 0.384 (0.023, 0.548) | 0.072 (0.019, 0.095) | 0.874 (0.778, 0.989) | 0.982 (0.975, 0.986) | 0.694 (0.615, 0.723) |
| LSTM | 60 | 0.077 (0.049, 0.104) | 0.117 (0.0, 0.175) | 0.336 (0.0, 0.524) | 0.083 (0.0, 0.194) | 0.89 (0.788, 0.999) | 0.981 (0.975, 0.985) | 0.696 (0.609, 0.74) |
| LSTM | 30 | 0.071 (0.044, 0.091) | 0.102 (0.0, 0.163) | 0.294 (0.0, 0.524) | 0.08 (0.0, 0.18) | 0.897 (0.743, 1.0) | 0.98 (0.975, 0.985) | 0.69 (0.614, 0.734) |
| LSTM | 15 | 0.067 (0.044, 0.093) | 0.101 (0.0, 0.16) | 0.282 (0.0, 0.548) | 0.074 (0.0, 0.135) | 0.9 (0.751, 0.997) | 0.98 (0.975, 0.985) | 0.683 (0.603, 0.727) |
|
| ||||||||
| Algorithm | Length of Timestep | AUPRC | F-Score | Sensitivity | PPV | Specificity | NPV | AUROC |
|
| ||||||||
| Time-to-outcome Dataset | ||||||||
| GRU | 60 | 0.101 (0.06, 0.137) | 0.142 (0.068, 0.203) | 0.443 (0.083, 0.619) | 0.092 (0.059, 0.175) | 0.873 (0.766, 0.982) | 0.984 (0.976, 0.988) | 0.717 (0.649, 0.752) |
| GRU | 30 | 0.087 (0.045, 0.117) | 0.137 (0.012, 0.184) | 0.421 (0.011, 0.608) | 0.088 (0.013, 0.138) | 0.884 (0.784, 0.992) | 0.984 (0.975, 0.988) | 0.714 (0.626, 0.754) |
| GRU | 15 | 0.086 (0.047, 0.116) | 0.139 (0.0, 0.188) | 0.459 (0.0, 0.608) | 0.084 (0.0, 0.114) | 0.875 (0.787, 0.993) | 0.985 (0.975, 0.988) | 0.722 (0.615, 0.766) |
| LSTM | 60 | 0.084 (0.048, 0.114) | 0.121 (0.0, 0.185) | 0.396 (0.0, 0.56) | 0.076 (0.0, 0.141) | 0.868 (0.771, 0.997) | 0.983 (0.975, 0.986) | 0.7 (0.61, 0.746) |
| LSTM | 30 | 0.085 (0.062, 0.109) | 0.128 (0.036, 0.186) | 0.408 (0.024, 0.584) | 0.088 (0.053, 0.161) | 0.875 (0.756, 0.992) | 0.983 (0.975, 0.986) | 0.708 (0.663, 0.747) |
| LSTM | 15 | 0.086 (0.055, 0.112) | 0.127 (0.0, 0.188) | 0.413 (0.0, 0.608) | 0.08 (0.0, 0.135) | 0.88 (0.8, 0.999) | 0.983 (0.975, 0.988) | 0.706 (0.635, 0.746) |
|
| ||||||||
| Algorithm | Length of Timestep | AUPRC | F-Score | Sensitivity | PPV | Specificity | NPV | AUROC |
|
| ||||||||
| Dataset of Single Datapoint | ||||||||
| Logistic Regression | N/A | 0.093 (0.09, 0.096) | 0.14 (0.134, 0.144) | 0.451 (0.429, 0.452) | 0.083 (0.079, 0.085) | 0.872 (0.866, 0.876) | 0.984 (0.984, 0.984) | 0.718 (0.712, 0.724) |
| Random Forest | N/A | 0.064 (0.055, 0.073) | 0.128 (0.103, 0.153) | 0.276 (0.214, 0.333) | 0.083 (0.067, 0.1) | 0.922 (0.914, 0.93) | 0.98 (0.979, 0.982) | 0.66 (0.628, 0.698) |
The GRU model has the best AUPRC of 0.101 (0.060, 0.137) and F-score of 0.142 (0,068, 0.203) on the time-to-outcome dataset of 60 minutes timestep.
Abbreviations: GRU, gated recurrent unit; LSTM, long short-term memory.
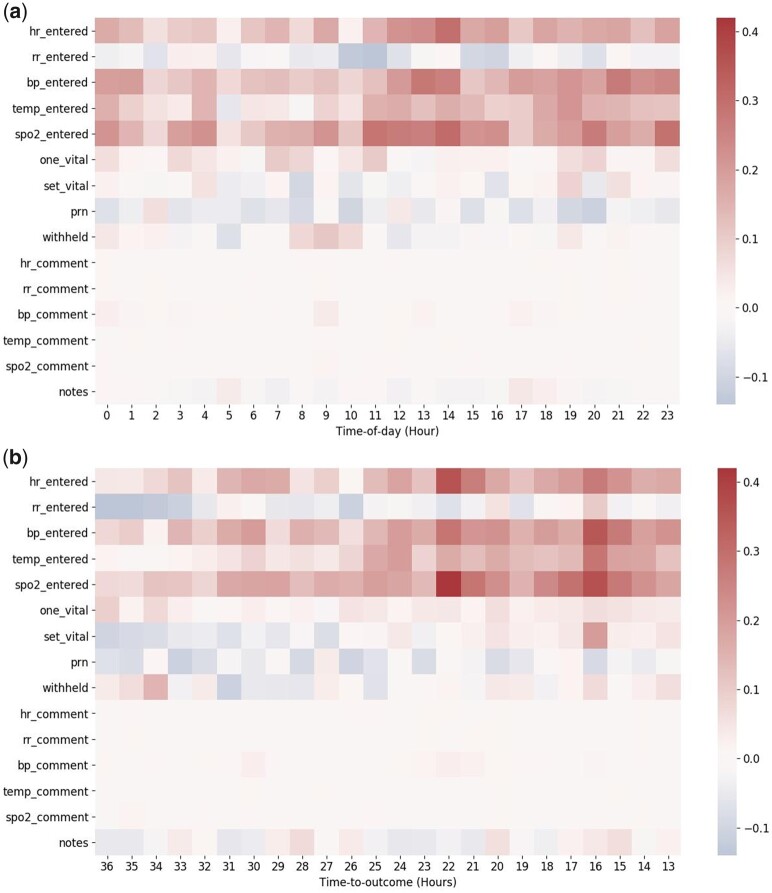
The values on the scale bar represent the difference between the average count of a given feature from ICU stays with outcomes and without outcomes. These averages are calculated for every feature per hour per patient. The redder the color for a particular feature, the more frequent that feature occurs in an ICU stay with outcome.
DISCUSSION
Our implementation of machine learning algorithms that use only timestamps from EHR metadata results in well-performing discriminative power. We emphasize that this approach doesn’t include the clinical measurement value (eg, heart rate = 90) but instead uses the frequency and co-occurrence of each data entry and documentation pattern that reflect clinicians’ practice patterns and clinical workflow. Additionally, our RNN algorithms display their abilities to learn temporal patterns, which results in improved discriminating power. This result supports our hypothesis that metadata in EHR, a proxy signal of the clinician expert’s objective and subjective assessments, provide valuable information on clinical outcomes, and thus have great potential to inform clinical prediction models.25,32,33,42
We curate the threshold for our best RNN model to achieve a discriminative power with sensitivity 0.443 and PPV 0.092 using the National Early Warning Score (0.395, 0.152) and Advanced Alert Monitoring (0.489,0.162) as benchmarks.40 The former is an expert opinion-based algorithm while the latter is a discrete-time logistic regression model derived from a database of 21 hospitals, and both have been in actual use in clinical settings. However, we are not able to directly compare our model against these benchmarks because those models require measurement values to compute and were validated on distinct study cohorts.
Clinical data is full of missing data and is irregularly measured. These characteristics have been a major challenge in longitudinal data analysis.43 Many EWS studies apply discrete time survival analysis on longitudinal data that completely exclude these characteristics as biases.10,40,44–46 Numerous RNN models incorporate information about missing data and irregularity as additional features.47–50 Our novel approach discretizes data into a multivariate time series representing the frequency and co-occurrence of clinical data entry. Consequently, we are able to preserve the irregularity characteristics as well as the missingness without adding additional artifacts and noise to the dataset. Finally, our results suggest that the dataset of timesteps of 60 minutes has sufficient granularity to represent the patterns of different clinical processes 36 hours before outcomes.
Time is an important component in clinical data because healthcare is not static.51 Observational measurements are often made when patients are ill. Clinicians make decisions based on their observations and previous understandings of their patients. Documentation of these observations, above and beyond regulatory requirements, are often made when clinicians are concerned about the result of a specific measurement or a patient’s clinical status.25,27 In the literature, the temporal pattern of lab orders also provides additional information on clinical outcomes regardless of the results.52,53 By exploiting time, we can capture this nonlinear recording process and positive feedback loop where measurement and health status affect each other.30,31,51 Our results have shown that time-to-outcome information further boosts RNN models’ AUPRC scores when compared to the baseline logistic regression model. Figure 3 demonstrates the difference of temporal patterns between ICU stays with outcomes and those without. The RNN models are able to learn these temporal patterns of different healthcare processes. We hypothesize that the time-of-day dataset provides additional information, indicative of records that happened in the rare hour of the day, which could further increase the model’s performance. However, our RNN models including the time-of-day information don’t achieve better performance. One explanation is that our method does embed the time-of-day information into the dataset but renders the original sequence of healthcare processes which affects our RNN models’ ability to recognize patients with higher risk.
Figure 3.
Comparison of the temporal patterns between ICU stays with outcomes and those without. The values on the scale bar represent the difference between the average count of a given feature from ICU stays with outcomes and without outcomes. These averages are calculated for every feature per hour per patient. The redder the color for a particular feature, the more frequent that feature occurs in an ICU stay with outcome. Figure 3a represents the relative signal intensity for time-of-day dataset. The x-axis indicates the time-of-day (hour) index. Figure 3b represents the relative signal intensity for time-to-outcome dataset. The x-axis indicates the time-to-outcome (hour) index.
Abbreviations: bp, blood pressure; hr, heart rate; one vital, single vital sign measurement; prn, one-time medication order; rr, respiratory rate; set vital, complete set of vital signs measurement; spo2, oxygen saturation; temp, body temperature; withhold, medication withhold.
There are several limitations in this study. Our analysis ran on a relatively small dataset from a healthcare system in Northeast US. This composite dataset includes various types of ICUs where the data represent different clinical processes and documentation patterns within the system. However, potential biases could derive from distinct patient population, healthcare process, regional practice, and, finally, individual clinician’s care pattern. Our results might not be generalizable to other healthcare systems. As a result, we plan to conduct further investigations on a larger dataset to verify the reproducibility of these findings and to externally validate our models. Secondly, our analysis only includes data from 36 to 12 hours before outcomes which limits our models’ ability to predict outcome in clinical practice. A future study will include model derivation on a forward-facing time window so that our models can be validated in a more realistic clinical setting. Moreover, our models don’t include measurement values that contain the information about the pathophysiological status of a patient. Therefore, we can’t directly compare our models with other well-established benchmark EWSs which require such values. A future study will include measurement values in order to validate our model against other benchmarks. Finally, our models are trained on timestamps of the underlying clinical data. There is often a delay when clinical data are entered into EHR which could lead to another bias if our models run on timestamps from EHR. However, our data preprocessing pipeline discretizes data into multivariate time series instead of using timestamps directly, which could potentially mitigate the bias created by the discrepancy between the timestamps of clinical data and EHR data. This discretization method also gives our model the potential to be implemented in real-time settings. Our models can generate predictions at any chosen timestep and requires no data imputation.
CONCLUSION
Clinicians’ recording and documentation patterns result in complex and diverse EHR data collection processes. This characteristic is seen by many as bias and is consequently overlooked in most clinical prediction modeling.21 Still, we believe it can also be exploited to yield valuable information for prediction model. This study demonstrates that our RNN models, using only timestamps of longitudinal EHR data that reflect healthcare processes, achieve well-performing discriminative power.
FUNDING
This work was funded by the National Institute for Nursing Research (NINR) funded CONCERN Study #1R01NR016941.
AUTHOR CONTRIBUTIONS
All listed authors substantially contributed to this work, including the study design, the acquisition, analysis, and interpretation of data. All authors participated in either drafting or revising this article and finally approved this version to be published.
DATA AVAILABILITY STATEMENT
The data underlying this article cannot be shared publicly due to inclusion of protected health information. The data will be shared on reasonable request to the corresponding author. All codes of our work are accessible on a public repository.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None decared.
Supplementary Material
REFERENCES
- 1.McQuillan P, Pilkington S, Allan A, et al. Confidential inquiry into quality of care before admission to intensive care. BMJ. Clin Res Ed. 1998; 316 (7148): 1853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaumont K, Luettel D, Thomson R.. Deterioration in hospital patients: early signs and appropriate actions. Nurs Stand (through 2013) 2008; 23 (1): 43–8. [DOI] [PubMed] [Google Scholar]
- 3.Hillman KM, Bristow PJ, Chey T, et al. Duration of life-threatening antecedents prior to intensive care admission. Intensive Care Med 2002; 28 (11): 1629–34. [DOI] [PubMed] [Google Scholar]
- 4.Bapoje SR, Gaudiani JL, Narayanan V, Albert RK.. Unplanned transfers to a medical intensive care unit: causes and relationship to preventable errors in care. J Hosp Med 2011; 6 (2): 68–72. [DOI] [PubMed] [Google Scholar]
- 5.Goldhill DR, White SA, Sumner A.. Physiological values and procedures in the 24 h before ICU admission from the ward. Anaesthesia 1999; 54 (6): 529–34. [DOI] [PubMed] [Google Scholar]
- 6.Schein RM, Hazday N, Pena M, Ruben BH, Sprung CL.. Clinical antecedents to in-hospital cardiopulmonary arrest. Chest 1990; 98 (6): 1388–92. [DOI] [PubMed] [Google Scholar]
- 7.Morgan RJMW, F, Wright MM.. An Early Warning Scoring System for detecting developing critical illness. Clin Intens Care 1997; 8: 100. [Google Scholar]
- 8.Prytherch DR, Smith GB, Schmidt PE, Featherstone PI.. ViEWS–Towards a national early warning score for detecting adult inpatient deterioration. Resuscitation 2010; 81 (8): 932–7. [DOI] [PubMed] [Google Scholar]
- 9.Subbe CP, Kruger M, Rutherford P, Gemmel L.. Validation of a modified Early Warning Score in medical admissions. QJM 2001; 94 (10): 521–6. [DOI] [PubMed] [Google Scholar]
- 10.Churpek MM, Yuen TC, Park SY, Gibbons R, Edelson DP.. Using electronic health record data to develop and validate a prediction model for adverse outcomes in the wards. Crit Care Med 2014; 42 (4): 841–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escobar GJ, LaGuardia JC, Turk BJ, Ragins A, Kipnis P, Draper D.. Early detection of impending physiologic deterioration among patients who are not in intensive care: development of predictive models using data from an automated electronic medical record. J Hosp Med 2012; 7 (5): 388–95. [DOI] [PubMed] [Google Scholar]
- 12.Churpek MM, Yuen TC, Park SY, Meltzer DO, Hall JB, Edelson DP.. Derivation of a cardiac arrest prediction model using ward vital signs. Crit Care Med 2012; 40 (7): 2102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Physicians RCo. National Early Warning Score (NEWS): Standardising the Assessment of Acute Illness Severity in the NHS. London: RCP; 2012. [Google Scholar]
- 14.Devita MA, Bellomo R, Hillman K, et al. Findings of the first consensus conference on medical emergency teams. Crit Care Med 2006; 34 (9): 2463–78. [DOI] [PubMed] [Google Scholar]
- 15.Jones DA, DeVita MA, Bellomo R.. Rapid-response teams. N Engl J Med 2011; 365 (2): 139–46. [DOI] [PubMed] [Google Scholar]
- 16.Moon A, Cosgrove JF, Lea D, Fairs A, Cressey DM.. An eight-year audit before and after the introduction of modified early warning score (MEWS) charts, of patients admitted to a tertiary referral intensive care unit after CPR. Resuscitation 2011; 82 (2): 150–4. [DOI] [PubMed] [Google Scholar]
- 17.Solomon RS, Corwin GS, Barclay DC, Quddusi SF, Dannenberg MD.. Effectiveness of rapid response teams on rates of in-hospital cardiopulmonary arrest and mortality: a systematic review and meta-analysis. J Hosp Med 2016; 11 (6): 438–45. [DOI] [PubMed] [Google Scholar]
- 18.Maharaj R, Raffaele I, Wendon J.. Rapid response systems: a systematic review and meta-analysis. Crit Care 2015; 19 (1): 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teuma Custo R, Trapani J.. The impact of rapid response systems on mortality and cardiac arrests: a literature review. Intensive Crit Care Nurs 2020; 59: 102848. [DOI] [PubMed] [Google Scholar]
- 20.Smith ME, Chiovaro JC, O'Neil M, et al. Early warning system scores for clinical deterioration in hospitalized patients: a systematic review. Ann Am Thorac Soc 2014; 11 (9): 1454–65. [DOI] [PubMed] [Google Scholar]
- 21.Fu LH, Schwartz J, Moy A, et al. Development and validation of early warning score system: a systematic literature review. J Biomed Inform 2020; 105: 103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothman MJ, Rothman SI, Beals J.. Development and validation of a continuous measure of patient condition using the electronic medical record. J Biomed Inform 2013; 46 (5): 837–48. [DOI] [PubMed] [Google Scholar]
- 23.Rapid Response Team composition, resourcing and calling criteria in Australia. Resuscitation 2012; 83 (5): 563–7. [DOI] [PubMed] [Google Scholar]
- 24.McGaughey J, O'Halloran P, Porter S, Blackwood B.. Early warning systems and rapid response to the deteriorating patient in hospital: A systematic realist review. J Adv Nurs 2017; 73 (12): 2877–91. [DOI] [PubMed] [Google Scholar]
- 25.Collins SA, Vawdrey DK. “Reading between the lines” of flow sheet data: nurses’ optional documentation associated with cardiac arrest outcomes. Appl Nurs Res 2012; 25 (4): 251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odell M, Victor C, Oliver D.. Nurses’ role in detecting deterioration in ward patients: systematic literature review. J Adv Nurs 2009; 65 (10): 1992–2006. [DOI] [PubMed] [Google Scholar]
- 27.Collins SA, Cato K, Albers D, et al. Relationship between nursing documentation and patients’ mortality.. Am J Crit Care 2013; 22 (4): 306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins SA, Fred M, Wilcox L, Vawdrey DK.. Workarounds used by nurses to overcome design constraints of electronic health records. In: proceedings of the 11th International Congress on Nursing Informatics; June 23–27, 2012; Montreal, Canada. [PMC free article] [PubMed] [Google Scholar]
- 29.Douw G, Schoonhoven L, Holwerda T, et al. Nurses’ worry or concern and early recognition of deteriorating patients on general wards in acute care hospitals: a systematic review. Crit Care 2015; 19 (1): 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013; 20 (1): 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hripcsak G, Albers DJ, Perotte A.. Exploiting time in electronic health record correlations. J Am Med Inform Assoc 2011; 18Suppl 1: i109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossetti SC, Knaplund C, Albers D, et al. Leveraging clinical expertise as a feature - not an outcome - of predictive models: evaluation of an early warning system use case. AMIA Annu Symp Proc 2019; 2019: 323–332. [PMC free article] [PubMed] [Google Scholar]
- 33.Rossetti SCK, Albers C, Dykes D, et al. Healthcare process modeling to phenotype clinician behaviors for exploiting the signal gain of clinical expertise (HPM-ExpertSignals): development and evaluation of a conceptual framework. J Am Med Inform Assoc 2021; 28 (6): 1242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:1412.3555; 2014.
- 35.Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 36.Tibshirani R.Regression shrinkage and selection via the Lasso. J Roy Stat Soc Ser B (Methodol) 1996; 58 (1): 267–88. [Google Scholar]
- 37.Hoerl AE, Kennard RW.. Ridge regression: biased estimation for nonorthogonal problems. Technometrics 1970; 12 (1): 55–67. [Google Scholar]
- 38.Li L, Jamieson K, DeSalvo G, Rostamizadeh A, Talwalkar A.. Hyperband: a novel bandit-based approach to hyperparameter optimization. J Mach Learn Res 2017; 18 (1): 6765–816. [Google Scholar]
- 39.Dziadzko MA, Novotny PJ, Sloan J, et al. Multicenter derivation and validation of an early warning score for acute respiratory failure or death in the hospital. Crit Care 2018; 22 (1): 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kipnis P, Turk BJ, Wulf DA, et al. Development and validation of an electronic medical record-based alert score for detection of inpatient deterioration outside the ICU. J Biomed Inform 2016; 64: 10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez CA, Clark CA, Zhang S, et al. Predicting out of intensive care unit cardiopulmonary arrest or death using electronic medical record data. BMC Med Inform Decis Mak 2013; 13: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins SA, Fred M, Wilcox L, Vawdrey DK.. Workarounds used by nurses to overcome design constraints of electronic health records. Ni 2012 (2012) 2012; 2012: 93. [PMC free article] [PubMed] [Google Scholar]
- 43.Ibrahim JG, Molenberghs G.. Missing data methods in longitudinal studies: a review. Test (Madr) 2009; 18 (1): 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Churpek MM, Yuen TC, Winslow C, Meltzer DO, Kattan MW, Edelson DP.. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med 2016; 44 (2): 368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Churpek MM, Adhikari R, Edelson DP.. The value of vital sign trends for detecting clinical deterioration on the wards. Resuscitation 2016; 102: 1–5. doi: 10.1016/j.resuscitation.2016.02.005[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Churpek MM, Yuen TC, Winslow C, et al. Multicenter development and validation of a risk stratification tool for ward patients. Am J Respir Crit Care Med 2014; 190 (6): 649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harutyunyan H, Khachatrian H, Kale DC, Ver Steeg G, Galstyan A.. Multitask learning and benchmarking with clinical time series data. Sci Data 2019; 6 (1): 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge W, Huh JW, Park YR, Lee JH, Kim YH, Turchin A.. An interpretable ICU mortality prediction model based on logistic regression and recurrent neural networks with LSTM units. AMIA Annu Symp Proc 2018; 2018: 460–9. [PMC free article] [PubMed] [Google Scholar]
- 49.Che Z, Purushotham S, Cho K, Sontag D, Liu Y.. Recurrent neural networks for multivariate time series with missing values. Sci Rep 2018; 8 (1): 6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipton ZC, Kale DC, Wetzel R. Modeling missing data in clinical time series with rnns. arXiv preprint arXiv:1606.04130; 2016.
- 51.Hripcsak G, Albers DJ.. High-fidelity phenotyping: richness and freedom from bias. J Am Med Inform Assoc 2018; 25 (3): 289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agniel D, Kohane IS, Weber GM.. Biases in electronic health record data due to processes within the healthcare system: retrospective observational study. BMJ (Clin Res Ed) 2018; 361: k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pivovarov R, Albers DJ, Sepulveda JL, Elhadad N.. Identifying and mitigating biases in EHR laboratory tests. J Biomed Inform 2014; 51: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to inclusion of protected health information. The data will be shared on reasonable request to the corresponding author. All codes of our work are accessible on a public repository.