Identification of the Candidate Proteins Related to Oleic Acid Accumulation during Peanut (Arachis hypogaea L.) Seed Development through Comparative Proteome Analysis
Abstract
:1. Introduction
2. Results
2.1. Morphology and Oil Accumulation of Developing Peanut Seeds
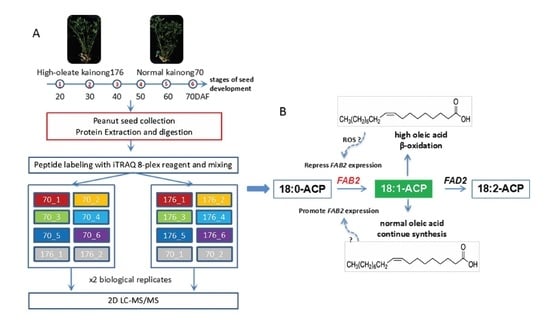
2.2. Mass Spectrometry Analysis and Protein Identification
2.3. Identification of DEPs (Differentially Expressed Proteins) during Seed Development in Low-Oleate Cultivar, Kainong70
2.4. Identification of DEPs during Seed Development in High-Oleate Cultivar Kainong176
2.5. Identification of DEPs at the Same Developmental Stage in Different Cultivars
2.6. Analysis of Expressions of DEPs at the mRNA Level
3. Discussion
4. Methods and Materials
4.1. Plant Materials
4.2. Profile Analysis of FAs
4.3. Protein Extraction and iTRAQ Labeling of Tryptic Peptides
4.4. LC-ESI-MS Analysis Based on Triple TOF 5600
4.5. Database Search
4.6. Total RNA Extraction and Real-Time PCR
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Akhtar, S.; Khalid, N.; Ahmed, I.; Shahzad, A.; Suleria, H.A. Physicochemical characteristics, functional properties, and nutritional benefits of peanut oil: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 1562–1575. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Qiu, J.; Agarwal, G.; Wang, J.; Ren, X.; Xia, H.; Guo, B.; Ma, C.; Wan, S.; Bertioli, D.J.; et al. Genome-Wide Discovery of Microsatellite Markers from Diploid Progenitor Species, Arachis duranensis and A. ipaensis, and Their Application in Cultivated Peanut (A. hypogaea). Front. Plant Sci. 2017, 8, 1209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, X.; Tian, L.; Chen, L.; Yu, W. Identification of peanut (Arachis hypogaea) chromosomes using a fluorescence in situ hybridization system reveals multiple hybridization events during tetraploid peanut formation. New Phytol. 2016, 211, 1424–1439. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, H.; Pandey, M.K.; Yang, Q.; Wang, X.; Garg, V.; Li, H.; Chi, X.; Doddamani, D.; Hong, Y.; et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. Proc. Natl. Acad. Sci. USA 2016, 113, 6785–6790. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cernay, C.; Pelzer, E.; Makowski, D. A global experimental dataset for assessing grain legume production. Sci. Data 2016, 3, 160084. [Google Scholar] [CrossRef] [PubMed]
- Vassiliou, E.K.; Gonzalez, A.; Garcia, C.; Tadros, J.H.; Chakraborty, G.; Toney, J.H. Oleic acid and peanut oil high in oleic acid reverse the inhibitory effect of insulin production of the inflammatory cytokine TNF-α both in vitro and in vivo systems. Lipids Health Dis. 2009, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Jung, S.; Moore, K.; Powell, G.; Ainsworth, C.; Abbott, A. High-oleate peanut mutants result from a MITE insertion into the FAD2 gene. Theor. Appl. Genet. 2004, 108, 1492–1502. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, X.; Zhang, X.; He, X.; Li, H.; Cui, D.; Yin, D. ITRAQ-Based Proteomic Analysis of the Metabolic Mechanisms behind Lipid Accumulation and Degradation during Peanut Seed Development and Postgermination. J. Proteome Res. 2016, 15, 4277–4289. [Google Scholar] [CrossRef] [PubMed]
- Li, H.F.; Zhu, F.H.; Li, H.Y.; Zhu, W.; Chen, X.P.; Hong, Y.B.; Liu, H.Y.; Wu, H.; Liang, X.Q. Proteomic identification of gravitropic response genes in peanut gynophores. J. Proteom. 2013, 93, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhao, S.; Hou, L.; Xia, H.; Wang, J.; Li, C.; Li, A.; Li, T.; Zhang, X.; Wang, X. Proteomics analysis reveals differentially activated pathways that operate in peanut gynophores at different developmental stages. BMC Plant Biol. 2015, 15, 188. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Dong, R.; Liu, L.; Hu, Z.; Li, J.; Wang, Y.; Ding, X.; Chu, Z. A novel mutant allele of SSI2 confers a better balance between disease resistance and plant growth inhibition on Arabidopsis thaliana. BMC Plant Biol. 2016, 16, 208. [Google Scholar] [CrossRef] [PubMed]
- Mekhedov, S.; de Ilarduya, O.M.; Ohlrogge, J. Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 2000, 122, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.S.; LaBrie, S.T.; Kinney, A.J.; von Wettstein-Knowles, P.; Browse, J. A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. 2002, 29, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Vellosillo, T.; Martinez, M.; Lopez, M.A.; Vicente, J.; Cascón, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.M.; Fulda, M.S.; Browse, J.A. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 2002, 129, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, F.; Gable, K.; Sayanova, O.; Dunn, T.; Napier, J.A. A Saccharomyces cerevisiae gene required for heterologous fatty acid elongase activity encodes a microsomal β-keto-reductase. J. Biol. Chem. 2002, 277, 11481–11488. [Google Scholar] [CrossRef] [PubMed]
- Branen, J.K.; Shintani, D.K.; Engeseth, N.J. Expression of antisense acyl carrier protein-4 reduces lipid content in Arabidopsis leaf tissue. Plant Physiol. 2003, 132, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Wen, T.N.; Nikolau, B.J.; Wurtele, E.S. Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme A carboxylase. Plant Physiol. 2000, 122, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Froman, B.E.; Edwards, P.C.; Bursch, A.G.; Dehesh, K. ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol. 2000, 123, 733–742. [Google Scholar] [CrossRef] [PubMed]
- De Boer, G.J.; Testerink, C.; Pielage, G.; Nijkamp, H.J.; Stuitje, A.R. Sequences surrounding the transcription initiation site of the Arabidopsis enoyl-acyl carrier protein reductase gene control seed expression in transgenic tobacco. Plant Mol. Biol. 1999, 39, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Katavic, V.; Li, F.; Haughn, G.W.; Kunst, L. Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. Plant J. 2010, 64, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Jessen, D.; Roth, C.; Wiermer, M.; Fulda, M. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol. 2015, 167, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef] [PubMed]
- Dehesh, K.; Tai, H.; Edwards, P.; Byrne, J.; Jaworski, J.G. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Z.; Xue, H.W. Arabidopsis beta-ketoacyl-[acyl carrier protein] synthase is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 2010, 22, 3726–3744. [Google Scholar] [CrossRef] [PubMed]
- Beld, J.; Lee, D.J.; Burkart, M.D. Fatty acid biosynthesis revisited: Structure elucidation and metabolic engineering. Mol. Biosyst. 2015, 11, 38–59. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2007, 63, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Block, M.A.; Jouhet, J. Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites. Curr. Opin. Cell Biol. 2015, 35, 21–29. [Google Scholar] [CrossRef] [PubMed]
- ALJohani, A.M.; Syed, D.N.; Ntambi, J.M. Insights into Stearoyl-CoA Desaturase-1 Regulation of Systemic Metabolism. Trends Endocrinol. Metab. 2017, 28, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Yang, Q.; Pan, L.; Chen, M.; He, Y.; Yang, Z.; Yu, S. Isolation and characterization of fatty acid desaturase genes from peanut (Arachis hypogaea L.). Plant Cell Rep. 2011, 30, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Bowen, K.J.; Kris-Etherton, P.M.; Shearer, G.C.; West, S.G.; Reddivari, L.; Jones, P. Oleic acid-derived oleoylethanolamide: A nutritional science perspective. Prog. Lipid Res. 2017, 67, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lung, S.C.; Chye, M.L. Acyl-CoA-Binding Proteins (ACBPs) in Plant Development. Subcell. Biochem. 2016, 86, 363–404. [Google Scholar] [PubMed]
- Mooney, B.P.; Miernyk, J.A.; Randall, D.D. The complex fate of alpha-ketoacids. Ann. Rev. Plant Biol. 2002, 53, 357–375. [Google Scholar] [CrossRef] [PubMed]
- LeClere, S.; Rampey, R.A.; Bartel, B. IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol. 2004, 135, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol. 2004, 134, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Kayam, G.; Faigenboim-Doron, A.; Clevenger, J.; Ozias-Akins, P.; Hovav, R. Gene expression profiling during seed-filling process in peanut with emphasis on oil biosynthesis networks. Plant Sci. 2016, 248, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Fan, S.; Wei, H.; Tao, C.; Ma, Q.; Ma, Q.; Zhang, S.; Li, H.; Pang, C.; Yu, S. iTRAQ-Based Quantitative Proteomic Analysis Reveals Cold Responsive Proteins Involved in Leaf Senescence in Upland Cotton (Gossypium hirsutum L.). Int. J. Mol. Sci. 2017, 18, 1984. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, S.; Gu, F.; Liu, W.; Yang, G.; Huang, M.; Xiao, W.; Liu, Y.; Guo, T.; Wang, H.; et al. NBS-LRR Protein Pik-H4 Interacts with OsBIHD1 to Balance Rice Blast Resistance and Growth by Coordinating Ethylene-Brassinosteroid Pathway. Front Plant Sci. 2017, 8, 127. [Google Scholar] [CrossRef] [PubMed]





| ID | Log2 Fold Change | Uniprot | Homologous Gene in Arabidopsis | Biological Process |
|---|---|---|---|---|
| kainong70-2 vs. Kainong70-1 | ||||
| Araip.10018346.1 | 2.67 | P52410 | KAS1 | fatty acid biosynthetic process |
| Araip.10018212.1 | 2.54 | Q9LX13 | At5g10160 | fatty acid biosynthetic process |
| Araip.10000461.1 | 2.13 | P49243 | At1g62640 | fatty acid biosynthetic process |
| Araip.10008089.1 | 2.12 | Q9C9P4 | KAS2 | unsaturated fatty acid metabolic process |
| Araip.10012310.1 | 1.97 | Q9SLA8 | MOD1 | fatty acid biosynthetic process |
| Araip.10010555.1 | 1.83 | Q9ZRA2 | HGO | lipid oxidation |
| Araip.10003244.1 | 1.8 | Q9SJH7 | CSY3 | lipid oxidation |
| Araip.10039800.1 | 1.8 | Q8RU07 | EMB3147 | fatty acid biosynthetic process |
| Araip.10007633.1 | 1.79 | Q06327 | LOX1 | fatty acid biosynthetic process |
| Araip.10037915.1 | 1.68 | Q9SQI8 | LTA2 | lipid biosynthetic process |
| Araip.10038396.1 | 1.66 | Q9FLW9 | PKP2 | lipid biosynthetic process |
| Araip.10036992.1 | 1.63 | Q9CAP8 | LACS9 | fatty acid metabolic process |
| Araip.10020945.1 | 1.61 | Q9SW21 | ACP4 | fatty acid biosynthetic process |
| Araip.10019224.1 | 1.47 | O82399 | PMDH1 | lipid oxidation |
| Araip.10019122.1 | 1.46 | F4JML5 | At4g16800 | lipid oxidation |
| Araip.10033415.1 | 1.44 | Q9ZP05 | PMDH2 | regulation of lipid catabolic process |
| Araip.10019397.1 | 1.27 | Q9LD43 | CAC3 | fatty acid biosynthetic process |
| Araip.10008554.1 | 1.27 | Q9M8L4 | GLPK | lipid oxidation |
| Araip.10034912.1 | 1.27 | Q9FLH8 | At5g51830 | lipid biosynthetic process |
| Araip.10032969.1 | 1.23 | Q9ZVQ3 | GSTZ1 | lipid oxidation |
| Araip.10039882.1 | 1.17 | Q8L9C4 | KCR1 | fatty acid biosynthetic process |
| Araip.10034519.1 | 1.08 | O04983 | CAC2 | fatty acid derivative biosynthetic process |
| Araip.10005278.1 | 1.03 | O04420 | At2g26230 | lipid oxidation |
| Araip.10021913.1 | 1.01 | Q9FMN0 | SCP2 | lipid oxidation |
| Araip.10000513.1 | −1.04 | Q38882 | PLDALPHA1 | fatty acid metabolic process |
| Araip.10003126.1 | −1.04 | O22898 | LACS1 | fatty acid metabolic process |
| Araip.10030492.1 | −1.05 | Q9T0A0 | LACS4 | fatty acid biosynthetic process |
| Araip.10004872.1 | −1.2 | O65202 | ACX1 | lipid oxidation |
| Araip.10013985.1 | −1.29 | Q9SCY5 | KINB2 | fatty acid biosynthetic process |
| Araip.10025239.1 | −1.49 | Q8RWZ3 | IBR3 | fatty acid metabolic process |
| Araip.10021376.1 | −1.52 | Q9FKE9 | At5g45160 | lipid oxidation |
| Araip.10022760.1 | −1.56 | Q9LZ31 | CYP77A4 | lipid oxidation |
| Araip.10000675.1 | −1.74 | Q8LCU7 | At3g45770 | fatty acid biosynthetic process |
| kainong70-3 vs. Kainong70-2 | ||||
| Araip.10019609.1 | 1.52 | Q93W03 | At3g56130 | negative regulation of fatty acid metabolic process |
| Araip.10037854.1 | 1.44 | Q42134 | PAE2 | lipid oxidation |
| Araip.10032974.1 | −1.11 | P42742 | PBF1 | lipid oxidation |
| Araip.10005498.1 | −1.25 | Q9SJH7 | CSY3 | lipid oxidation |
| Araip.10020218.1 | −1.34 | Q96329 | ACX4 | lipid oxidation |
| Araip.10008534.1 | −1.46 | Q94FY7 | VTE1 | fat-soluble vitamin metabolic process |
| Araip.10030492.1 | −1.6 | Q9T0A0 | LACS4 | fatty acid biosynthetic process |
| Araip.10036992.1 | −1.64 | Q9CAP8 | LACS9 | fatty acid metabolic process |
| Araip.10014023.1 | −2.07 | Q9ZPI5 | MFP2 | lipid oxidation |
| Araip.10038212.1 | −2.14 | Q8LPS1 | LACS6 | fatty acid biosynthetic process |
| Kainong70-4 vs. Kainong70-3 | ||||
| Araip.10021376.1 | 1.57 | Q9FKE9 | At5g45160 | lipid oxidation |
| Araip.10020945.1 | −1.67 | Q9SW21 | ACP4 | fatty acid biosynthetic process |
| Kainong70-5 vs. Kainong70-4 | ||||
| no identified | ||||
| Kainong70-5 vs. Kainong70-4 | ||||
| Araip.10029908.1 | 1.87 | Q9M9W8 | PLPZETA2 | lipid oxidation |
| Araip.10020945.1 | 1.79 | Q9SW21 | ACP4 | fatty acid biosynthetic process |
| Araip.10010590.1 | 1.45 | Q9LDF5 | At3g15290 | fatty acid metabolic process |
| Araip.10020381.1 | 1.08 | O22832 | FAB2 | fatty acid biosynthetic process |
| ID | Log2 Fold Change | Uniprot | Homologous Gene in Arabidopsis | Biological Process |
|---|---|---|---|---|
| Kainong176-2 vs. Kainong176-1 | ||||
| Araip.10018212.1 | 2.52 | Q9LX13 | At5g10160 | fatty acid biosynthetic process |
| Araip.10018346.1 | 2.49 | P52410 | KAS1 | fatty acid biosynthetic process |
| Araip.10000461.1 | 2.4 | P49243 | At1g62640 | fatty acid biosynthetic process |
| Araip.10012310.1 | 2.24 | Q9SLA8 | MOD1 | fatty acid biosynthetic process |
| Araip.10019224.1 | 2.11 | O82399 | PMDH1 | lipid oxidation |
| Araip.10003244.1 | 1.86 | Q9SJH7 | CSY3 | lipid oxidation |
| Araip.10039800.1 | 1.86 | Q8RU07 | EMB3147 | fatty acid biosynthetic process |
| Araip.10038396.1 | 1.85 | Q9FLW9 | PKP2 | lipid biosynthetic process |
| Araip.10038248.1 | 1.85 | Q9LUJ7 | PAP85 | lipid storage |
| Araip.10007633.1 | 1.81 | Q06327 | LOX1 | fatty acid biosynthetic process |
| Araip.10033415.1 | 1.78 | Q9ZP05 | PMDH2 | regulation of fatty acid oxidation |
| Araip.10008554.1 | 1.75 | Q9M8L4 | GLPK | lipid oxidation |
| Araip.10010555.1 | 1.74 | Q9ZRA2 | HGO | lipid oxidation |
| Araip.10008089.1 | 1.71 | Q9C9P4 | KAS2 | lipid biosynthetic process |
| Araip.10039818.1 | 1.71 | P33207 | At1g24360 | fatty acid biosynthetic process |
| Araip.10037915.1 | 1.67 | Q9SQI8 | LTA2 | lipid biosynthetic process |
| Araip.10019122.1 | 1.64 | F4JML5 | At4g16800 | fatty acid metabolic process |
| Araip.10020945.1 | 1.62 | Q9SW21 | ACP4 | fatty acid biosynthetic process |
| Araip.10014644.1 | 1.62 | Q9SS98 | At3g01570 | lipid storage |
| Araip.10014038.1 | 1.34 | Q9M7Z1 | BCE2 | fatty acid biosynthetic process |
| Araip.10039882.1 | 1.33 | Q8L9C4 | KCR1 | fatty acid biosynthetic process |
| Araip.10004625.1 | 1.26 | Q9FVS9 | CYP96A15 | fatty acid derivative metabolic process |
| Araip.10034912.1 | 1.25 | Q9FLH8 | At5g51830 | lipid metabolic process |
| Araip.10032969.1 | 1.23 | Q9ZVQ3 | GSTZ1 | lipid oxidation |
| Araip.10034519.1 | 1.13 | O04983 | CAC2 | lipid biosynthetic process |
| Araip.10000107.1 | 1.02 | P56765 | accD | lipid biosynthetic process |
| Araip.10036992.1 | 1 | Q9CAP8 | LACS9 | fatty acid metabolic process |
| Araip.10004872.1 | −1.04 | O65202 | ACX1 | lipid oxidation |
| Araip.10003126.1 | −1.11 | O22898 | LACS1 | fatty acid metabolic process |
| Araip.10034349.1 | −1.25 | Q9ZPI6 | AIM1 | lipid oxidation |
| Araip.10003587.1 | −1.3 | Q9FFE6 | AAE5 | fatty acid metabolic process |
| Araip.10023165.1 | −1.42 | P0CZ23 | ACX3 | lipid oxidation |
| Araip.10017616.1 | −1.49 | O82265 | SCC3 | lipid oxidation |
| Araip.10000675.1 | −1.55 | Q8LCU7 | At3g45770 | fatty acid biosynthetic process |
| Araip.10025239.1 | −1.71 | Q8RWZ3 | IBR3 | fatty acid metabolic process |
| Kainong176-3 vs. Kainong176-2 | ||||
| Araip.10027909.1 | 2.74 | Q93Y35 | RPN7 | lipid oxidation |
| Araip.10018302.1 | 2.3 | Q9SGW3 | RPN12A | lipid oxidation |
| Araip.10021150.1 | 2.2 | Q06588 | ACO4 | lipid oxidation |
| Araip.10020381.1 | 1.97 | O22832 | FAB2 | fatty acid biosynthetic process |
| Araip.10019397.1 | 1.96 | Q9LD43 | CAC3 | fatty acid biosynthetic process |
| Araip.10039882.1 | 1.95 | Q8L9C4 | KCR1 | fatty acid biosynthetic process |
| Araip.10003887.1 | 1.91 | O80992 | PYL2 | fatty acid metabolic process |
| Araip.10032969.1 | 1.89 | Q9ZVQ3 | GSTZ1 | lipid oxidation |
| Araip.10031916.1 | 1.79 | Q9LLC1 | BCCP2 | fatty acid biosynthetic process |
| Araip.10031864.1 | 1.69 | Q38997 | KIN10 | lipid biosynthetic process |
| Araip.10004243.1 | 1.48 | Q9FLW9 | PKP2 | lipid biosynthetic process |
| Araip.10018529.1 | 1.46 | Q9ZRW8 | GSTU19 | lipid oxidation |
| Araip.10019122.1 | 1.44 | F4JML5 | At4g16800 | fatty acid beta-oxidation |
| Araip.10035390.1 | 1.42 | Q9SEI4 | RPT3 | lipid oxidation |
| Araip.10000267.1 | 1.4 | Q9M2U2 | ECR | fatty acid biosynthetic process |
| Araip.10037915.1 | 1.38 | Q9SQI8 | LTA2 | lipid biosynthetic process |
| Araip.10020218.1 | 1.33 | Q96329 | ACX4 | lipid oxidation |
| Araip.10006248.1 | 1.32 | Q9S9W2 | SDRA | fatty acid metabolic process |
| Araip.10034583.1 | 1.29 | Q9SS98 | At3g01570 | lipid storage |
| Araip.10034519.1 | 1.18 | O04983 | CAC2 | lipid biosynthetic process |
| Araip.10007455.1 | 1.16 | Q8GRT9 | At3g15690 | cellular lipid metabolic process |
| Araip.10028882.1 | 1.15 | Q96242 | CYP74A | fatty acid derivative metabolic process |
| Araip.10039818.1 | 1.09 | P33207 | At1g24360 | fatty acid biosynthetic process |
| Araip.10007632.1 | 1.01 | Q06327 | LOX1 | fatty acid biosynthetic process |
| Araip.10036036.1 | −1.05 | Q9C5U1 | AHK3 | response to lipid |
| Araip.10020945.1 | −1.11 | Q9SW21 | ACP4 | fatty acid biosynthetic process |
| Araip.10018212.1 | −1.24 | Q9LX13 | At5g10160 | fatty acid biosynthetic process |
| Araip.10024168.1 | −1.25 | Q8LBB2 | KING1 | fatty acid biosynthetic process |
| Araip.10014023.1 | −1.29 | Q9ZPI5 | MFP2 | lipid oxidation |
| Araip.10027878.1 | −1.39 | Q9LST0 | At5g60160 | lipid oxidation |
| Araip.10018115.1 | −1.49 | Q8VXZ7 | AGAL3 | lipid oxidation |
| Araip.10039592.1 | −1.52 | Q9SIE3 | At2g22230 | lipid biosynthetic process |
| Araip.10034411.1 | −1.61 | Q7DLS1 | PBB2 | lipid oxidation |
| Araip.10013255.1 | −1.63 | O65201 | ACX2 | lipid oxidation |
| Araip.10023663.1 | −1.69 | Q9LT08 | RPN11 | lipid oxidation |
| Araip.10010590.1 | −1.77 | Q9LDF5 | At3g15290 | fatty acid metabolic process |
| Araip.10030656.1 | −1.79 | Q8GYB8 | OPR2 | fatty acid biosynthetic process |
| Araip.10024291.1 | −1.85 | B9DGD6 | ACS | fatty acid biosynthetic process |
| Araip.10029015.1 | −1.94 | Q944G9 | FBA2 | lipid biosynthetic process |
| Araip.10013985.1 | −2.29 | Q9SCY5 | KINB2 | fatty acid biosynthetic process |
| Araip.10010555.1 | −2.33 | Q9ZRA2 | HGO | lipid oxidation |
| Araip.10001867.1 | −2.51 | Q8S4Y1 | AAT1 | cellular lipid catabolic process |
| Kainong176-4 vs. Kainong176-3 | ||||
| Araip.10028103.1 | 2.39 | Q9FKE9 | At5g45160 | lipid oxidation |
| Araip.10023165.1 | 1.5 | P0CZ23 | ACX3 | lipid oxidation |
| Araip.10037915.1 | −1.1 | Q9SQI8 | LTA2 | unsaturated fatty acid biosynthetic process |
| Araip.10030536.1 | −1.13 | F4HUK6 | AAE1 | fatty acid metabolic process |
| Araip.10003887.1 | −1.37 | O80992 | PYL2 | fatty acid metabolic process |
| Araip.10032969.1 | −1.57 | Q9ZVQ3 | GSTZ1 | lipid oxidation |
| Araip.10019122.1 | −1.63 | F4JML5 | At4g16800 | fatty acid metabolic process |
| Araip.10039818.1 | −2.14 | P33207 | At1g24360 | fatty acid metabolic process |
| Kainong176-5 vs. Kainong176-4 | ||||
| no identified | ||||
| Kainong176-6 vs. Kainong176-5 | ||||
| Araip.10015142.1 | −2.02 | Q9M8L4 | GLPK | lipid oxidation |
| ID | Log2 Fold Change | Uniprot | Homologous Gene in Arabidopsis | Biological Process | Seed Development Stage |
|---|---|---|---|---|---|
| Araip.10001989.1 | −1.5 | Q9FJ62 | GDPDL4 | lipid metabolic process | 1 |
| Araip.10013985.1 | 1.34 | Q9SCY5 | KINB2 | fatty acid biosynthetic | 2 |
| Araip.10032497.1 | −1.03 | Q9C826 | ABA2 | lipid metabolic process | 2 |
| Araip.10039818.1 | 2.03 | P33207 | At1g24360 | fatty acid biosynthetic | 3 |
| Araip.10020381.1 | 1.82 | O22832 | FAB2 | fatty acid biosynthetic | 3 |
| Araip.10019122.1 | 1.62 | F4JML5 | At4g16800 | fatty acid metabolic process | 3 |
| Araip.10012320.1 | 1.54 | Q9SYT0 | ANN1 | fatty acid metabolic process | 3 |
| Araip.10037915.1 | 1.37 | Q9SQI8 | LTA2 | fatty acid metabolic process | 3 |
| Araip.10021150.1 | 2.06 | Q06588 | ACO4 | lipid metabolic process | 3 |
| Araip.10007105.1 | 1.42 | O64688 | E1-BETA-2 | lipid metabolic process | 3 |
| Araip.10033916.1 | 1.23 | P0DKC6 | HSD1 | lipid metabolic process | 3 |
| Araip.10037384.1 | 1.11 | Q9C8P0 | EMB3003 | carbohydrate metabolic process | 3 |
| Araip.10026147.1 | −1.34 | F4JQJ7 | At4g36945 | lipid biosynthetic | 3 |
| Araip.10023165.1 | −1.64 | P0CZ23 | ACX3 | lipid oxidation | 3 |
| Araip.10006287.1 | −1.64 | O65390 | APA1 | lipid metabolic process | 3 |
| Araip.10033474.1 | −1.72 | F4J7G5 | At3g11780 | lipid metabolic process | 3 |
| Araip.10015601.1 | −2.51 | Q9M153 | At4g01130 | GDSL-like Lipase | 3 |
| Araip.10040083.1 | 1.31 | Q38862 | IPS2 | lipid Inositol-3-phosphate synthase | 4 |
| Araip.10017616.1 | 1.03 | O82265 | SCC3 | lipid oxidation | 4 |
| Araip.10003529.1 | −2 | Q9ZVI9 | PECT1 | phospholipid metabolic process | 4 |
| Araip.10026823.1 | −2.15 | Q9LDB4 | LTP6 | non-specific lipid-transfer protein 10-related | 4 |
| Araip.10040083.1 | 1.66 | Q38862 | IPS2 | lipid Inositol-3-phosphate synthase | 5 |
| Araip.10010555.1 | 2.07 | Q9ZRA2 | HGO | lipid oxidation | 6 |
| Araip.10024204.1 | 1.53 | Q8L7U0 | At3g03330 | lipid oxidation | 6 |
| Araip.10006678.1 | 1.35 | Q9FMA3 | PEX5 | lipid oxidation | 6 |
| Araip.10033474.1 | 1.97 | F4J7G5 | At3g11780 | lipid metabolic process | 6 |
| Araip.10020381.1 | −1.18 | O22832 | FAB2 | fatty acid biosynthetic | 6 |
| Araip.10005278.1 | −1.51 | O04420 | At2g26230 | lipid oxidation | 6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Li, H.; Gu, J.; Deng, L.; Ren, L.; Hong, Y.; Lu, Q.; Chen, X.; Liang, X. Identification of the Candidate Proteins Related to Oleic Acid Accumulation during Peanut (Arachis hypogaea L.) Seed Development through Comparative Proteome Analysis. Int. J. Mol. Sci. 2018, 19, 1235. https://doi.org/10.3390/ijms19041235
Liu H, Li H, Gu J, Deng L, Ren L, Hong Y, Lu Q, Chen X, Liang X. Identification of the Candidate Proteins Related to Oleic Acid Accumulation during Peanut (Arachis hypogaea L.) Seed Development through Comparative Proteome Analysis. International Journal of Molecular Sciences. 2018; 19(4):1235. https://doi.org/10.3390/ijms19041235
Chicago/Turabian StyleLiu, Hao, Haifen Li, Jianzhong Gu, Li Deng, Li Ren, Yanbin Hong, Qing Lu, Xiaoping Chen, and Xuanqiang Liang. 2018. "Identification of the Candidate Proteins Related to Oleic Acid Accumulation during Peanut (Arachis hypogaea L.) Seed Development through Comparative Proteome Analysis" International Journal of Molecular Sciences 19, no. 4: 1235. https://doi.org/10.3390/ijms19041235
APA StyleLiu, H., Li, H., Gu, J., Deng, L., Ren, L., Hong, Y., Lu, Q., Chen, X., & Liang, X. (2018). Identification of the Candidate Proteins Related to Oleic Acid Accumulation during Peanut (Arachis hypogaea L.) Seed Development through Comparative Proteome Analysis. International Journal of Molecular Sciences, 19(4), 1235. https://doi.org/10.3390/ijms19041235