Abstract
RNA interference (RNAi) refers to the ability of exogenously introduced double-stranded RNA to silence expression of homologous sequences. Silencing is initiated when the enzyme Dicer processes the double-stranded RNA into small interfering RNAs (siRNAs). Small RNA molecules are incorporated into Argonaute-protein-containing effector complexes, which they guide to complementary _targets to mediate different types of gene silencing, specifically post-transcriptional gene silencing and chromatin-dependent gene silencing1. Although endogenous small RNAs have crucial roles in chromatin-mediated processes across kingdoms, efforts to initiate chromatin modifications in trans by using siRNAs have been inherently difficult to achieve in all eukaryotic cells. Using fission yeast, here we show that RNAi-directed heterochromatin formation is negatively controlled by the highly conserved RNA polymerase-associated factor 1 complex (Paf1C). Temporary expression of a synthetic hairpin RNA in Paf1C mutants triggers stable heterochromatin formation at homologous loci, effectively silencing genes in trans. This repressed state is propagated across generations by the continual production of secondary siRNAs, independently of the synthetic hairpin RNA. Our data support a model in which Paf1C prevents _targeting of nascent transcripts by the siRNA-containing RNA-induced transcriptional silencing complex and thereby epigenetic gene silencing, by promoting efficient transcription termination and rapid release of the RNA from the site of transcription. We show that although compromised transcription termination is sufficient to initiate the formation of bi-stable heterochromatin by trans-acting siRNAs, impairment of both transcription termination and nascent transcript release is imperative to confer stability to the repressed state. Our work uncovers a novel mechanism for small-RNA-mediated epigenome regulation and highlights fundamental roles for Paf1C and the RNAi machinery in building epigenetic memory.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 (2009)
Volpe, T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002)
Verdel, A. et al. RNAi-mediated _targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004)
Motamedi, M. R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004)
Schalch, T. et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol. Cell 34, 36–46 (2009)
Castel, S. E. & Martienssen, R. A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nature Rev. Genet. 14, 100–112 (2013)
Chan, S. W., Zhang, X., Bernatavichute, Y. V. & Jacobsen, S. E. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 4, e363 (2006)
Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A. & Matzke, A. J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201 (2000)
Ting, A. H., Schuebel, K. E., Herman, J. G. & Baylin, S. B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nature Genet. 37, 906–910 (2005)
Morris, K. V., Chan, S. W., Jacobsen, S. E. & Looney, D. J. Small interfering rna-induced transcriptional gene silencing in human cells. Science 305, 1289–1292 (2004)
Kim, D. H., Villeneuve, L. M., Morris, K. V. & Rossi, J. J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nature Struct. Mol. Biol. 13, 793–797 (2006)
Janowski, B. A. et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nature Struct. Mol. Biol. 13, 787–792 (2006)
Napoli, S., Pastori, C., Magistri, M., Carbone, G. M. & Catapano, C. V. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 28, 1708–1719 (2009)
Simmer, F. et al. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep. 11, 112–118 (2010)
Iida, T., Nakayama, J. & Moazed, D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell 31, 178–189 (2008)
Mbogning, J. et al. The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet. 9, e1004029 (2013)
Yu, R., Jih, G., Iglesias, N. & Moazed, D. Determinants of heterochromatic siRNA biogenesis and function. Mol. Cell 53, 262–276 (2014)
Bühler, M., Verdel, A. & Moazed, D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125, 873–886 (2006)
Tomson, B. N. & Arndt, K. M. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829, 116–126 (2013)
Cheung, A. C. & Cramer, P. A movie of RNA polymerase II transcription. Cell 149, 1431–1437 (2012)
Aranda, A. & Proudfoot, N. Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell 7, 1003–1011 (2001)
Herr, A. J., Molnar, A., Jones, A. & Baulcombe, D. C. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl Acad. Sci. USA 103, 14994–15001 (2006)
Halic, M. & Moazed, D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140, 504–516 (2010)
Ptashne, M. On the use of the word ‘epigenetic’. Curr. Biol. 17, R233–R236 (2007)
Chandler, V. L. Paramutation: from maize to mice. Cell 128, 641–645 (2007)
Luteijn, M. J. & Ketting, R. F. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nature Rev. Genet. 14, 523–534 (2013)
Luteijn, M. J. et al. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430 (2012)
Chandler, V. L. Paramutation’s properties and puzzles. Science 330, 628–629 (2010)
Ding, L. et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4, 403–415 (2009)
Ponnusamy, M. P. et al. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells 27, 3001–3011 (2009)
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene _targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998)
Emmerth, S. et al. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev. Cell 18, 102–113 (2010)
Keller, C. et al. HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol. Cell 47, 215–227 (2012)
Leeds, P., Peltz, S. W., Jacobson, A. & Culbertson, M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5, 2303–2314 (1991)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498 (2011)
Ihaka, R. & Gentleman, R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 (1996)
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004)
Huber, W., Toedling, J. & Steinmetz, L. M. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics 22, 1963–1970 (2006)
Bühler, M., Haas, W., Gygi, S. P. & Moazed, D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129, 707–721 (2007)
Acknowledgements
We thank T. Iida for providing the plasmid encoding the ade6-hp construct, N. Laschet and R. Tsuji for technical assistance, S. Thiry for hybridizing tiling arrays, K. Jacobeit and S. Dessus-Babus for small RNA sequencing, T. Roloff for archiving data sets, M. Kirschmann for developing the Matlab script for colony counting, and A. Tuck for comments on the manuscript. This work was supported by funds from the Swiss National Science Foundation, the European Research Council, and the Boehringer Ingelheim Fonds. The Friedrich Miescher Institute for Biomedical Research is supported by the Novartis Research Foundation.
Author information
Authors and Affiliations
Contributions
Y.S., K.M.K., V.F. and J.B. generated strains and performed experiments; Y.S. performed the sms screen; the genome-wide small RNA and gene expression data were analysed by K.M.K.; M.B.S. designed and performed the computational analysis of the mutant genome resequencing data; M.B. designed experiments and prepared the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed.
Extended data figures and tables
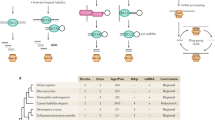
Extended Data Figure 1 Design of the ade6+ RNA hairpin (ade6-hp) construct that expresses abundant sense and antisense (primary) siRNAs.
a, The RNA stem–loop construct consists of a 250-nucleotide-long ade6+ fragment, followed by a cox4+ intronic sequence and the reverse complement of the ade6+ fragment. The promoter sequence of the adh1+ gene drives expression of the RNA hairpin. Transcription of the construct is terminated by the termination signals of the nmt1+ gene. The construct was provided by T. Iida. b, c, Small RNA sequencing revealed that the RNA stem is converted into sense and antisense siRNAs covering the 250-nucleotide stretch from the ade6+ open reading frame (nucleotides 621–870). Furthermore, sense and antisense siRNAs mapping to the cox4+ intronic and adh1+ promoter sequences are also generated when this construct is expressed in wild-type cells. ORF, open reading frame. Asterisk denotes the point mutation (Thr645Ala) in the ade6-704 loss of function allele. Green arrows indicate forward and reverse primers that were used for PCR in ChIP experiments. d, Schematic diagram depicting origin and _target(s) of synthetic ade6-hp siRNAs. The ade6-hp expression cassette (a) was inserted into the nmt1+ locus on chromosome I by homologous recombination. The ade6-hp-containing plasmid was linearized with PmlI, which cuts in the middle of the nmt1+ terminator sequence, and transformed into ade6-704 cells. Thereby, the ade6-hp construct was inserted downstream of the nmt1+ gene. The nourseothricin (Nat)-resistance cassette linked to the ade6-hp construct allowed selection of positive transformants. It also allows assessment of spreading of repressive heterochromatin that is nucleated by the ade6-hp siRNAs in cis (see Extended Data Fig. 7b). A wild-type copy of the ade6+ gene was inserted upstream of the trp1+ gene on chromosome II by homologous recombination. Because the endogenous ade6-704 allele is non-functional, positive transformants could be selected by growth in the absence of adenine. In Paf1C mutant cells, ade6-hp-derived siRNAs either act in cis to assemble heterochromatin at the nmt1+ locus (chromosome I), or in trans to direct the formation of heterochromatin at the trp1+::ade6+ (chromosome II) and ade6-704 (chromosome III) loci.
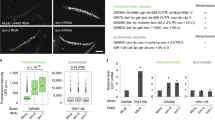
Extended Data Figure 2 Silencing assays demonstrating the inability of synthetic siRNAs to act in trans in Paf1C wild-type cells.
a, ade6+ silencing assays were performed with cells expressing synthetic ade6-hp siRNAs, ura4-hp siRNAs or no siRNAs. The ability of ade6-hp siRNAs to silence either the endogenous ade6+ gene or the trp1+::ade6+ reporter gene was assessed at different adenine concentrations. ade6-704 cells were used as positive control. b, c, ade6+ mRNA levels were determined by quantitative RT–PCR and normalized to act1+ mRNA. One representative biological replicate is shown. Error bars, s.d. d, ade6+silencing assays demonstrating that neither the endogenous ade6+ gene nor the trp1+::ade6+ reporter gene becomes repressed by trans-acting ade6-hp siRNAs, even upon overexpression of the heterochromatin protein Swi6.
Extended Data Figure 3 Sms forward genetic screen identifies five true positive hits that enable siRNAs to methylate H3K9 at the ade6+ gene in trans.
a, Workflow of the EMS mutagenesis screen. We mutagenized sms0 cells, which express abundant siRNAs complementary to the ade6+ gene (indicated by green hairpin), with EMS (primary screen). Subsequently, we tested the positive red colonies for growth in the absence of adenine to select against loss-of-function mutations in the adenine biosynthesis pathway (secondary screen). In hits that remained positive after the secondary screen, dcr1+ was deleted to identify truly siRNA-dependent hits (tertiary screen). For mapping of causative mutations by whole-genome next-generation sequencing, positive hits were backcrossed four times. b, sms1-10 mutants show the red ade6+ silencing phenotype on YE plates, which segregated through four successive backcrosses for all 10 mutants. The ade6-M210 loss-of-function allele and ade6+ inserted within centromeric heterochromatin (otr1R::ade6+) serve as positive controls. c, ChIP experiment demonstrating methylation of H3K9 at the ade6+ _target loci in sms1, 3, 4, 6 and 8. One representative biological replicate is shown. d, ade6+ silencing in sms1, 3, 4, 6 and 8 is Dcr1-dependent. e, Resequencing of EMS-mutagenized S. pombe strains. From outside to inside, the tracks show the genomic location, the average coverage per window of 10 kb (black line, scale from 0 to 30), the number of sequence variations identified before filtering in all strains per window of 10 kb (blue bars, scale from 0 to 90) and the five mutations that passed the filtering and overlapped with Paf1C genes (red lines, the two mutations in Paf1 are too close to be resolved individually). f, Table lists mutations mapped by whole-genome sequencing. In Dcr1-dependent mutants, we mapped mutations in the genes SPBC651.09c, SPAC664.03, SPBC13E7.08c and SPBC17G9.02c whose homologues in budding yeast encode for protein subunits of the Paf1 complex.
Extended Data Figure 4 Mutant alleles for the homologues of all five subunits of Paf1C enable siRNAs to induce gene silencing in trans.
a, ade6+ siRNAs reduce ade6+ mRNA levels in all Paf1C mutant strains identified in this study. Whole-genome tiling arrays were used to assess gene expression in the mutant cells indicated. y axis is in linear scale. b, C-terminally tagged Tpr1 and Cdc73 are hypomorphic. Full deletions of the tpr1+ and cdc73+ genes cause retarded growth phenotypes (Extended Data Fig. 8c). By contrast, tpr1-3xFLAG and cdc73-3xFLAG grow normally, and display ade6-hp siRNA-mediated repression of the ade6+ gene.
Extended Data Figure 5 Expression of synthetic siRNAs in paf1-Q264Stop cells is sufficient to trigger stable repression of protein coding genes in trans.
a, Left, the paf1-Q264Stop mutation was introduced into cells that express synthetic ura4-hp siRNAs15. Right, wild-type (paf1+) and paf1-Q264Stop were grown in the presence or absence of 5-FOA. Red arrow indicates paf1-Q264Stop colonies growing on FOA-containing medium. Note that these colonies could be propagated in non-selective medium without losing the repressed state. b, In S. pombe, artificial tethering of the RITS complex to mRNA expressed from the endogenous ura4+ locus using the phage λN protein results in de novo generation of ura4+ siRNAs. These siRNAs load onto RITS and are necessary to establish heterochromatin at the ura4+ locus in cis. However, like ura4-hp siRNAs, they are incapable of triggering the repression of a second ura4+ locus in trans18. To test whether ura4+ siRNAs produced as a result of Tas3λN tethering to ura4+::5BoxB mRNA (chromosome III) can act in trans to silence a second ura4+ allele (leu1Δ::ura4+, chromosome II), paf1+ was mutated and ura4+ repression was assessed by FOA silencing assays. Whereas 5-FOA was toxic to both paf1+ and paf1-Q264Stop cells in the absence of ura4+ siRNAs (Tas3 not fused to λN), FOA-resistant colonies appeared upon Tas3-λN tethering, demonstrating that siRNAs generated from the ura4+::5BoxB locus can initiate repression of the second ura4+ copy expressed from the leu1+ locus. Notably, siRNA-mediated ura4+ repression in trans was more pronounced in the absence of the RNase Eri1. We have previously shown that the levels of ura4+::5BoxB-derived siRNA are higher in eri1Δ cells41. We note that trans-silencing of the second ura4+ allele occasionally occurs in paf1+ cells in the absence of Eri1 (ref. 18). However, in contrast to paf1-Q264Stop cells, the repressed state of ura4+ is not stably propagated. Hairpin symbols downstream of the ura4+ ORF denote BoxB sequences. They form stem–loop structures when transcribed and are bound by the λN protein. c, ade6+ silencing assay demonstrating that also the endogenous ade6+ gene is repressed if ade6-hp siRNAs are expressed from the nmt1+ locus in paf1-Q264Stop cells. Silencing assay was performed with two freshly generated (naive) paf1-Q264Stop mutant strains. A few white colonies in which heterochromatin has not yet formed are discernable. Such white colonies were picked to determine heterochromatin initiation frequencies shown in Fig. 2.
Extended Data Figure 6 ade6+ siRNAs trigger de novo methylation of H3K9 at homologous ade6+ sequences in cis and in trans.
a, ade6-hp RNA producing locus and siRNA _target loci in trans in the sms0 strain. ade6-704 is a loss-of-function allele of the endogenous ade6+ gene and serves as a positive control in the silencing assays. b, c, ade6+ siRNAs direct the methylation of H3K9 at ade6 _targets in cis (green) and in trans (red) in Paf1C mutant cells. H3K9me2 for trp1+::ade6+ is shown in Fig. 1d. Quantitative PCR was performed with locus-specific primers. Error bars, s.e.m.; n = 3 technical replicates. d, e, ChIP experiments to assess ade6+ transcriptional activity. H3K36me3 levels were normalized to total H3 levels. snu6+ is transcribed by RNAPIII and serves as background control. Error bars, s.e.m.; n = 3 independent biological replicates; P values were calculated using the one-tailed Student’s t-test.
Extended Data Figure 7 Pronounced siRNA-directed heterochromatin formation in trans during meiosis.
a, White (naive) cells that had not yet established heterochromatin at the trp1+::ade6+ locus were isolated from populations of freshly generated paf1-Q264Stop strains and crossed with paf1+ cells. Both mating partners expressed ade6-hp siRNAs and contained the same trp1+::ade6+ reporter. Spores were dissected on YE plates and incubated for 3–4 days at 30 °C. Note the non-Mendelian inheritance pattern of the parental white phenotype and the high incidence of heterochromatin formation (red phenotype) in paf1-Q264Stop cells after meiosis. b, Spores from 43 tetrads were dissected in total. Colonies formed by the individual spores (a) were then struck on YE plates and incubated for 3–4 days at 30 °C, followed by replica-plating onto YES-G418 and YES+nourseothricin (Nat) plates for genotyping. Thus, the cells visible on the YE plates have gone through roughly 50–80 mitotic divisions after mating and sporulation. This analysis shows that de novo formation of heterochromatin by trans-acting siRNAs during meiosis occurs more frequently than in mitosis. However, once established, heterochromatin is remarkably stable in mitotic cells (see also Fig. 2). Notably, growth of some paf1-Q264Stop descendants was reduced on YES+Nat plates, demonstrating spreading of heterochromatin into the neighbouring Nat-resistance cassette that marks the nmt1+::ade6-hp+ locus (see Extended Data Fig. 1). Note that genes repressed by heterochromatin can be derepressed under strong negative selection. Thus, this observation indicates extraordinary repressive activity of the heterochromatin that forms in cis at the ade6-hp siRNA-producing locus. Finally, paf1+ cells (no growth on YES-G418 or PMG-LEU) never turned red, demonstrating the high repressive activity of Paf1. This explains unsatisfactory results of previous attempts to induce the formation of stable heterochromatin in trans by expressing synthetic siRNAs.
Extended Data Figure 8 Effect of Paf1C mutations on global gene expression and silencing.
a, The effect of the Paf1C mutations on genome expression was assessed by hybridizing total RNA to whole-genome tiling arrays. The parental wild-type strain, all Paf1C point mutations discovered in the screen, and full deletions of the paf1+ and leo1+ genes were included in the analysis. To compare the genome-wide expression profiles of the mutants with the wild-type strain, a principal component analysis (PCA) was performed on the data obtained for two biological replicates of each strain. Principal component (PC) 1 and 2 explained 41.5% and 16.4% of the variance between samples and were selected for visualization, revealing that cdc73-G313R and paf1Δ cells are most different from wild-type cells. All the other mutants clustered together in a group of samples that also includes wild type, demonstrating that RNA steady-state levels are only minimally affected in these mutants. Note that leo1Δ is more similar to wild type than paf1Δ, as well as that paf1Δ clusters separately from the Paf1C point mutants. b, Pairwise comparisons of gene expression between wild-type and paf1 mutant strains. c, leo1Δ cells have no growth defect but are susceptible for de novo formation of heterochromatin by siRNAs acting in trans. These results suggest that Leo1 might be a bona fide repressor of small-RNA-mediated heterochromatin formation.
Extended Data Figure 9 Kinetic model for Paf1C-mediated repression of siRNA-directed heterochromatin formation.
a, Paf1C facilitates rapid transcription and release of the nascent transcript from the DNA template. Because the kinetics of transcription termination and RNA 3′ end processing is faster than RITS binding and CLRC recruitment, stable heterochromatin and long-lasting gene silencing cannot be established. b, In Paf1C mutant cells identified in this study, elongation of RNA polymerase II, termination of transcription, and the release of the nascent transcript from the site of transcription is decelerated. This results in an accumulation of RNA polymerases that are associated with nascent transcripts, opening up a window of opportunity for the siRNA-guided RITS complex to base-pair with nascent transcripts and recruit CLRC. Consequently, highly stable and repressive heterochromatin is assembled, which is accompanied by the generation of secondary siRNAs covering the entire locus (not depicted in this scheme). Notably, our results demonstrate that impaired transcription termination but not elongation is sufficient to allow silencing. However, to confer robustness to the repressed state, both transcription termination and release of the RNA transcript from the site of transcription must be impaired concomitantly.
Extended Data Figure 10 Formation of ectopic heterochromatin.
a, Differential gene expression compared to differential antisense siRNA expression in leo1Δ. Gene expression profiles were obtained with whole-genome tiling arrays and small RNA profiles by deep sequencing. Genes neighbouring the nmt1+::ade6-hp+, trp1+::ade6+ and ade6-704 loci are marked in colour (see also Supplementary Table 1). b, siRNA reads mapping to the ade6-704 locus in leo1+ and leo1Δ strains. Red, plus strand; blue, minus strand. Intronic rpl2302 siRNAs in leo1Δ cells indicate co-transcriptional double-stranded RNA synthesis by RDRC before splicing. c, ChIP experiment showing H3K9me2 enrichments on genes surrounding the ade6-704 locus in leo1+ and leo1Δ cells. Enrichments were calculated relative to background levels obtained in clr4Δ cells and normalized to adh1+. Error bars, s.d.; mean of n = 2 independent biological replicates.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4. (PDF 272 kb)
Rights and permissions
About this article
Cite this article
Kowalik, K., Shimada, Y., Flury, V. et al. The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature 520, 248–252 (2015). https://doi.org/10.1038/nature14337
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14337
This article is cited by
-
The dynamics and functional mechanisms of H2B mono-ubiquitination
Crop Health (2024)
-
Polymeric nature of tandemly repeated genes enhances assembly of constitutive heterochromatin in fission yeast
Communications Biology (2023)
-
A cis-acting mechanism mediates transcriptional memory at Polycomb _target genes in mammals
Nature Genetics (2021)
-
Mating can initiate stable RNA silencing that overcomes epigenetic recovery
Nature Communications (2021)
-
Flowering and flowering genes: from model plants to orchids
Horticulture, Environment, and Biotechnology (2021)