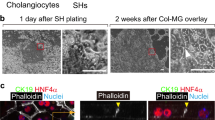
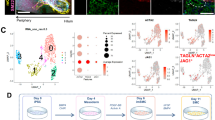
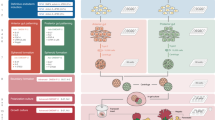
Abstract
The treatment of common bile duct (CBD) disorders, such as biliary atresia or ischemic strictures, is restricted by the lack of biliary tissue from healthy donors suitable for surgical reconstruction. Here we report a new method for the isolation and propagation of human cholangiocytes from the extrahepatic biliary tree in the form of extrahepatic cholangiocyte organoids (ECOs) for regenerative medicine applications. The resulting ECOs closely resemble primary cholangiocytes in terms of their transcriptomic profile and functional properties. We explore the regenerative potential of these organoids in vivo and demonstrate that ECOs self-organize into bile duct–like tubes expressing biliary markers following transplantation under the kidney capsule of immunocompromised mice. In addition, when seeded on biodegradable scaffolds, ECOs form tissue-like structures retaining biliary characteristics. The resulting bioengineered tissue can reconstruct the gallbladder wall and repair the biliary epithelium following transplantation into a mouse model of injury. Furthermore, bioengineered artificial ducts can replace the native CBD, with no evidence of cholestasis or occlusion of the lumen. In conclusion, ECOs can successfully reconstruct the biliary tree, providing proof of principle for organ regeneration using human primary cholangiocytes expanded in vitro.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Murray, K.F. & Carithers, R.L. Jr. AASLD practice guidelines: evaluation of the patient for liver transplantation. Hepatology 41, 1407–1432 (2005).
Perkins, J.D. Are we reporting the same thing? Liver Transpl. 13, 465–466 (2007).
Skaro, A.I. et al. The impact of ischemic cholangiopathy in liver transplantation using donors after cardiac death: the untold story. Surgery 146, 543–552, discussion 552–553 (2009).
Enestvedt, C.K. et al. Biliary complications adversely affect patient and graft survival after liver retransplantation. Liver Transpl. 19, 965–972 (2013).
Gallo, A. & Esquivel, C.O. Current options for management of biliary atresia. Pediatr. Transplant. 17, 95–98 (2013).
Felder, S.I. et al. Hepaticojejunostomy using short-limb Roux-en-Y reconstruction. JAMA Surg. 148, 253–257, discussion 257–258 (2013).
Sampaziotis, F., Segeritz, C.-P. & Vallier, L. Potential of human induced pluripotent stem cells in studies of liver disease. Hepatology 62, 303–311 (2015).
Kanno, N., LeSage, G., Glaser, S., Alvaro, D. & Alpini, G. Functional heterogeneity of the intrahepatic biliary epithelium. Hepatology 31, 555–561 (2000).
Sampaziotis, F. et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 33, 845–852 (2015).
Sampaziotis, F. et al. Directed differentiation of human induced pluripotent stem cells into functional cholangiocyte-like cells. Nat. Protoc. 12, 814–827 (2017).
LeSage, G., Glaser, S. & Alpini, G. Regulation of cholangiocyte proliferation. Liver 21, 73–80 (2001).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Masyuk, A.I., Masyuk, T.V. & LaRusso, N.F. Cholangiocyte primary cilia in liver health and disease. Dev. Dyn. 237, 2007–2012 (2008).
Tabibian, J.H., Masyuk, A.I., Masyuk, T.V., O'Hara, S.P. & LaRusso, N.F. Physiology of cholangiocytes. Compr. Physiol. 3, 541–565 (2013).
Cízková, D., Morký, J., Micuda, S., Osterreicher, J. & Martínková, J. Expression of MRP2 and MDR1 transporters and other hepatic markers in rat and human liver and in WRL 68 cell line. Physiol. Res. 54, 419–428 (2005).
Gigliozzi, A. et al. Molecular identification and functional characterization of Mdr1a in rat cholangiocytes. Gastroenterology 119, 1113–1122 (2000).
Xia, X., Francis, H., Glaser, S., Alpini, G. & LeSage, G. Bile acid interactions with cholangiocytes. World J. Gastroenterol. 12, 3553–3563 (2006).
Caperna, T.J., Blomberg, A., Garrett, W.M. & Talbot, N.C. Culture of porcine hepatocytes or bile duct epithelial cells by inductive serum-free media. In Vitro Cell. Dev. Biol. Anim. 47, 218–233 (2011).
Marinelli, R.A. et al. Secretin induces the apical insertion of aquaporin-1 water channels in rat cholangiocytes. Am. J. Physiol. 276, G280–G286 (1999).
Gong, A.-Y. et al. Somatostatin stimulates ductal bile absorption and inhibits ductal bile secretion in mice via SSTR2 on cholangiocytes. Am. J. Physiol. Cell Physiol. 284, C1205–C1214 (2003).
Ito, M. et al. NOD/SCID/γcnull mouse: an excellent recipient mouse model for engraftment of human cells. Blood 100, 3175–3182 (2002).
Chan, B.P. & Leong, K.W. Scaffolding in tissue engineering: general approaches and tissue-specific considerations. Eur. Spine J. 17 (Suppl. 4), 467–479 (2008).
Cheung, H.-Y., Lau, K.-T., Lu, T.-P. & Hui, D. A critical review on polymer-based bio-engineered materials for scaffold development. Compos., Part B Eng. 38, 291–300 (2007).
Jabłonska, B. End-to-end ductal anastomosis in biliary reconstruction: indications and limitations. Can. J. Surg. 57, 271–277 (2014).
Gjorevski, N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016).
Koo, B.K. & Clevers, H. Stem cells marked by the R-spondin receptor LGR5. Gastroenterology 147, 289–302 (2014).
Farin, H.F., Van Es, J.H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529. e7 (2012).
Du, P., Kibbe, W.A. & Lin, S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Lin, S.M., Du, P., Huber, W. & Kibbe, W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36, e11 (2008).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Bertero, A. et al. Activin/nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes Dev. 29, 702–717 (2015).
Campos, P.B., Sartore, R.C., Abdalla, S.N. & Rehen, S.K. Chromosomal spread preparation of human embryonic stem cells for karyotyping. J. Vis. Exp. http://dx.doi.org/10.3791/1512 (2009).
Hannan, N.R.F. et al. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports 1, 293–306 (2013).
Koo, B.-K., Sasselli, V. & Clevers, H. Retroviral gene expression control in primary organoid cultures. Curr. Protoc. Stem Cell Biol. 27, Unit 5A.6 (2013).
Shultz, L.D. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 174, 6477–6489 (2005).
Acknowledgements
The authors would like to thank J. Skepper, L. Carter and the University of Cambridge Advanced Imaging Centre for their help with electron microscopy; E. Farnell and the University of Cambridge, Cambridge Genomic Services for their help with microarray data processing and analysis; A. Petrunkina and the NIHR Cambridge BRC Cell Phenotyping Hub for their help with cell sorting; K. Burling and the MRC MDU Mouse Biochemistry Laboratory (MRC_MC_UU_12012/5) for processing mouse serum samples; and R. El-Khairi for her help with IF images, R. Grandy for his help with providing relevant references, the Cambridge Biorepository for Translational Medicine for the provision of human tissue used in the study; D. Trono (Ecole Polytechnique Federale de Lausanne) for the gift of the plasmids used for the generation of GFP-expressing ECOs and B. McLeod for IT support. The monoclonal antibody TROMA-III, developed by R. Kemler, was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa.
This work was funded by ERC starting grant Relieve IMDs (281335; L.V., N.R.F.H.), the Cambridge Hospitals National Institute for Health Research Biomedical Research Centre (L.V., N.R.F.H., S. Sinha., F.S.), the Evelyn Trust (N.H.) and the EU FP7 grant TissuGEN (M.C.D.B.) and was supported in part by the Intramural Research Program of the NIH/NIAID (R.L.G., C.A.R.). F.S. has been supported by an Addenbrooke's Charitable Trust Clinical Research Training Fellowship and a joint MRC–Sparks Clinical Research Training Fellowship. (MR/L016761/1) A.W.J. and A.E.M. acknowledge support from EPSRC (EP/L504920/1) and an Engineering for Clinical Practice Grant from the Department of Engineering, University of Cambridge. J.B. was supported by a BHF Studentship (Grant FS/13/65/30441).
Author information
Authors and Affiliations
Contributions
F.S. conceived and designed the study, performed experiments, acquired, interpreted and analyzed the data, developed and validated the protocols described, generated the figures, and wrote and edited the manuscript. A.W.J. generated the tubular densified collagen scaffolds and conceived and developed the manufacturing technique. O.C.T. contributed to cell culture and performed animal experiments, including kidney capsule injections and provision and dissection of mouse tissue. S. Sawiak performed the magnetic resonance imaging experiments. E.M.G. and S.S.U. reviewed and reported the magnetic resonance images. R.L.G. performed experiments, including animal experiments, IF and tissue histology. M.C.d.B. contributed to cell culture, generated viral particles, performed viral transduction experiments and generated GFP-expressing ECOs. N.L.B. and L. Valestrand performed animal experiments. M.J.G.-V. and P.M. performed bioinformatics analyses. D.O. performed flow cytometry analyses, and L.Y. performed immunoblot analyses. A.R. performed IF and qRT–PCR analyses and provided positive controls for IF and qRT–PCR. A.B. performed flow cytometry analyses and provided bioinformatics support. J.B. contributed to tissue histology and IF experiments. M.C.F.Z. contributed to PGA scaffold preparation and population with cells. M.T.P. generated viral particles, performed viral transduction experiments and generated GFP-expressing ECOs. M.P. generated viral particles. G.M.S. contributed to scaffold generation. P.M.M. and K.E.S. maintained and provided fibroblast controls. N.P. contributed to tissue culture. N.G. and C.A.R. contributed to dissection and provision of primary tissue. I.S. performed karyotyping and comparative genomic hybridization analyses. S.E.D. reviewed and reported the histology images. W.S., J.C., K.B.J., M.Z., S. Sinha, W.T.H.G., G.J.A., S.E.B., T.A.W., T.H.K. and E.M. contributed through critical revision of the manuscript for important intellectual content. N.R.F.H. contributed to the design and concept of the study and provided early study supervision. A.E.M. contributed to the design of the densified collagen scaffold and contributed through critical revision of the manuscript for important intellectual content. K.S.-P. provided primary tissue, performed animal experiments, including biliary reconstruction surgery, contributed to the design and concept of the study, supervised the study, interpreted the data, and wrote and edited the manuscript. L. Vallier designed and conceived the study, supervised the study, interpreted the data, and wrote and edited the manuscript. All of the authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
L. Vallier is a founder and shareholder of DefiniGEN. The remaining authors have nothing to disclose.
Supplementary information
Supplementary Data, Figures and Tables
Supplementary Figures 1–14 and Supplementary Tables 1 and 3–5 (PDF 3856 kb)
Supplementary Table 2
Microarray gene expression data corresponding to the heat map in Figure 1d. (XLSX 817 kb)
41591_2017_BFnm4360_MOESM3_ESM.mp4
Magnetic resonance cholangio-pancreatography (MRCP; sagital plane) of an extrahepatic biliary injury mouse 104 d after biliary reconstruction with an ECO-populated scaffold. (MP4 596 kb)
41591_2017_BFnm4360_MOESM4_ESM.mp4
T1-weighed magnetic resonance imaging (coronal plane) of an extrahepatic biliary injury mouse 104 d after biliary reconstruction with an ECO-populated scaffold. (MP4 2803 kb)
41591_2017_BFnm4360_MOESM5_ESM.mp4
T2-weighed magnetic resonance imaging (coronal plane) of an immunocompromised mouse 1 month after biliary reconstruction with an ECO-populated scaffold. (MP4 1128 kb)
Rights and permissions
About this article
Cite this article
Sampaziotis, F., Justin, A., Tysoe, O. et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat Med 23, 954–963 (2017). https://doi.org/10.1038/nm.4360
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4360
This article is cited by
-
Organoids and organoid extracellular vesicles-based disease treatment strategies
Journal of Nanobiotechnology (2024)
-
Patient-derived organoids in human cancer: a platform for fundamental research and precision medicine
Molecular Biomedicine (2024)
-
Organoid bioprinting: from cells to functional tissues
Nature Reviews Bioengineering (2024)
-
Stable two- and three-dimensional cholangiocyte culture systems from extrahepatic bile ducts of biliary atresia patients: use of structural and functional bile duct epithelium models for in vitro analyses
Cytotechnology (2024)
-
Progress, application and challenges of liver organoids
Clinical Cancer Bulletin (2024)