Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm
- PMID: 19717626
- PMCID: PMC2772452
- DOI: 10.1128/AEM.00974-09
Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm
Abstract
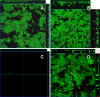
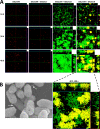
A conjugative plasmid from the catheter-associated urinary tract infection strain Escherichia coli MS2027 was sequenced and annotated. This 42,644-bp plasmid, designated pMAS2027, contains 58 putative genes and is most closely related to plasmids belonging to incompatibility group X (IncX1). Plasmid pMAS2027 encodes two important virulence factors: type 3 fimbriae and a type IV secretion (T4S) system. Type 3 fimbriae, recently found to be functionally expressed in E. coli, played an important role in biofilm formation. Biofilm formation by E. coli MS2027 was specifically due to expression of type 3 fimbriae and not the T4S system. The T4S system, however, accounted for the conjugative ability of pMAS2027 and enabled a non-biofilm-forming strain to grow as part of a mixed biofilm following acquisition of this plasmid. Thus, the importance of conjugation as a mechanism to spread biofilm determinants was demonstrated. Conjugation may represent an important mechanism by which type 3 fimbria genes are transferred among the Enterobacteriaceae that cause device-related infections in nosocomial settings.
Figures


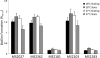
Similar articles
-
Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation.J Bacteriol. 2008 Feb;190(3):1054-63. doi: 10.1128/JB.01523-07. Epub 2007 Nov 30. J Bacteriol. 2008. PMID: 18055599 Free PMC article.
-
Molecular Epidemiology of Plasmid-Mediated Types 1 and 3 Fimbriae Associated with Biofilm Formation in Multidrug Resistant Escherichia coli from Diseased Food Animals in Guangdong, China.Microbiol Spectr. 2022 Oct 26;10(5):e0250321. doi: 10.1128/spectrum.02503-21. Epub 2022 Aug 15. Microbiol Spectr. 2022. PMID: 35969065 Free PMC article.
-
Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion.J Bacteriol. 2006 May;188(10):3582-8. doi: 10.1128/JB.188.10.3582-3588.2006. J Bacteriol. 2006. PMID: 16672612 Free PMC article.
-
The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model.Infect Immun. 2009 Jun;77(6):2272-84. doi: 10.1128/IAI.01333-08. Epub 2009 Mar 23. Infect Immun. 2009. PMID: 19307211 Free PMC article.
-
Bacterial biofilm formation, pathogenicity, diagnostics and control: An overview.Indian J Med Sci. 2009 Jul;63(7):313-21. Indian J Med Sci. 2009. PMID: 19700915 Review.
Cited by
-
Biofilm through the Looking Glass: A Microbial Food Safety Perspective.Pathogens. 2022 Mar 12;11(3):346. doi: 10.3390/pathogens11030346. Pathogens. 2022. PMID: 35335670 Free PMC article. Review.
-
The influence of biofilms in the biology of plasmids.Microbiol Spectr. 2014 Oct 10;2(5):0012. doi: 10.1128/microbiolspec.PLAS-0012-2013. Microbiol Spectr. 2014. PMID: 25392747 Free PMC article.
-
Characterization of nontyphoidal Salmonella strains from a tertiary hospital in China: serotype diversity, multidrug resistance, and genetic insights.Front Cell Infect Microbiol. 2024 Jan 9;13:1327092. doi: 10.3389/fcimb.2023.1327092. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38264733 Free PMC article.
-
Plasmid content of a clinically relevant Klebsiella pneumoniae clone from the Czech Republic producing CTX-M-15 and QnrB1.Antimicrob Agents Chemother. 2013 Feb;57(2):1073-6. doi: 10.1128/AAC.01886-12. Epub 2012 Dec 10. Antimicrob Agents Chemother. 2013. PMID: 23229477 Free PMC article.
-
Identification and characterization of microcin S, a new antibacterial peptide produced by probiotic Escherichia coli G3/10.PLoS One. 2012;7(3):e33351. doi: 10.1371/journal.pone.0033351. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479389 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Burmolle, M., M. L. Bahl, L. B. Jensen, S. J. Sorensen, and L. H. Hansen. 2008. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154:187-195. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

