miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway
- PMID: 25406066
- PMCID: PMC4235011
- DOI: 10.7554/eLife.01977
miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway
Abstract
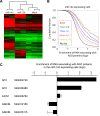
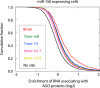
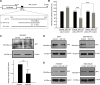
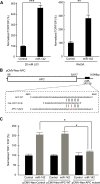
MicroRNAs (miRNAs) are important regulators of stem and progenitor cell functions. We previously reported that miR-142 and miR-150 are upregulated in human breast cancer stem cells (BCSCs) as compared to the non-tumorigenic breast cancer cells. In this study, we report that miR-142 efficiently recruits the APC mRNA to an RNA-induced silencing complex, activates the canonical WNT signaling pathway in an APC-suppression dependent manner, and activates the expression of miR-150. Enforced expression of miR-142 or miR-150 in normal mouse mammary stem cells resulted in the regeneration of hyperproliferative mammary glands in vivo. Knockdown of endogenous miR-142 effectively suppressed organoid formation by BCSCs and slowed tumor growth initiated by human BCSCs in vivo. These results suggest that in some tumors, miR-142 regulates the properties of BCSCs at least in part by activating the WNT signaling pathway and miR-150 expression.
Keywords: APC; WNT signaling pathway; breast cancer; cancer stem cells; cell biology; human; human biology; medicine; miR-142; miR-150; mouse.
Conflict of interest statement
KL: This author is a principal scientist of the Applied Biosystems.
MFC: Michael Clarke holds stock of the Oncomed Pharmaceuticals that focuses on development of therapeutic methods to _target cancer stem cells.
The other authors declare that no competing interests exist.
Figures
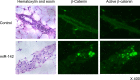
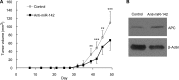
Similar articles
-
microRNA-128-3p overexpression inhibits breast cancer stem cell characteristics through suppression of Wnt signalling pathway by down-regulating NEK2.J Cell Mol Med. 2020 Jul;24(13):7353-7369. doi: 10.1111/jcmm.15317. Epub 2020 Jun 17. J Cell Mol Med. 2020. PMID: 32558224 Free PMC article.
-
Essential role of miR-200c in regulating self-renewal of breast cancer stem cells and their counterparts of mammary epithelium.BMC Cancer. 2015 Sep 23;15:645. doi: 10.1186/s12885-015-1655-5. BMC Cancer. 2015. PMID: 26400441 Free PMC article.
-
LncCCAT1 Promotes Breast Cancer Stem Cell Function through Activating WNT/β-catenin Signaling.Theranostics. 2019 Oct 1;9(24):7384-7402. doi: 10.7150/thno.37892. eCollection 2019. Theranostics. 2019. PMID: 31695775 Free PMC article.
-
The role of microRNAs in breast cancer stem cells.Int J Mol Sci. 2013 Jul 15;14(7):14712-23. doi: 10.3390/ijms140714712. Int J Mol Sci. 2013. PMID: 23860207 Free PMC article. Review.
-
MicroRNAs, a Promising _target for Breast Cancer Stem Cells.Mol Diagn Ther. 2020 Feb;24(1):69-83. doi: 10.1007/s40291-019-00439-5. Mol Diagn Ther. 2020. PMID: 31758333 Review.
Cited by
-
mTOR/miR-142-3p/PRAS40 signaling cascade is critical for tuberous sclerosis complex-associated renal cystogenesis.Cell Mol Biol Lett. 2024 Sep 27;29(1):125. doi: 10.1186/s11658-024-00638-x. Cell Mol Biol Lett. 2024. PMID: 39333852 Free PMC article.
-
Adipsin-dependent adipocyte maturation induces cancer cell invasion in breast cancer.Sci Rep. 2024 Aug 9;14(1):18494. doi: 10.1038/s41598-024-69476-3. Sci Rep. 2024. PMID: 39122742 Free PMC article.
-
Transplantation of Wnt5a-modified Bone Marrow Mesenchymal Stem Cells Promotes Recovery After Spinal Cord Injury via the PI3K/AKT Pathway.Mol Neurobiol. 2024 Dec;61(12):10830-10844. doi: 10.1007/s12035-024-04248-8. Epub 2024 May 25. Mol Neurobiol. 2024. PMID: 38795301 Free PMC article.
-
miRNA on the Battlefield of Cancer: Significance in Cancer Stem Cells, WNT Pathway, and Treatment.Cancers (Basel). 2024 Feb 27;16(5):957. doi: 10.3390/cancers16050957. Cancers (Basel). 2024. PMID: 38473318 Free PMC article. Review.
-
Unraveling the Potential of miRNAs from CSCs as an Emerging Clinical Tool for Breast Cancer Diagnosis and Prognosis.Int J Mol Sci. 2023 Nov 6;24(21):16010. doi: 10.3390/ijms242116010. Int J Mol Sci. 2023. PMID: 37958993 Free PMC article. Review.
References
-
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortes ML, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Piña V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio RC, Parkin M, Pho NH, Quintanar-Jurado V, Ramos AH, Rebollar-Vega R, Rodriguez-Cuevas S, Romero-Cordoba SL, Schumacher SE, Stransky N, Thompson KM, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi DC, Richardson AL, Jimenez-Sanchez G, Lander ES, Gabriel SB, Garraway LA, Golub TR, Melendez-Zajgla J, Toker A, Getz G, Hidalgo-Miranda A, Meyerson M. 2012. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486:405–409. doi: 10.1038/nature11154. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

