BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins
- PMID: 29788290
- PMCID: PMC6030817
- DOI: 10.1093/nar/gky383
BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins
Abstract
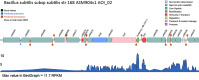
Interest in secondary metabolites such as RiPPs (ribosomally synthesized and posttranslationally modified peptides) is increasing worldwide. To facilitate the research in this field we have updated our mining web server. BAGEL4 is faster than its predecessor and is now fully independent from ORF-calling. Gene clusters of interest are discovered using the core-peptide database and/or through HMM motifs that are present in associated context genes. The databases used for mining have been updated and extended with literature references and links to UniProt and NCBI. Additionally, we have included automated promoter and terminator prediction and the option to upload RNA expression data, which can be displayed along with the identified clusters. Further improvements include the annotation of the context genes, which is now based on a fast blast against the prokaryote part of the UniRef90 database, and the improved web-BLAST feature that dynamically loads structural data such as internal cross-linking from UniProt. Overall BAGEL4 provides the user with more information through a user-friendly web-interface which simplifies data evaluation. BAGEL4 is freely accessible at http://bagel4.molgenrug.nl.
Figures

Similar articles
-
BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W448-53. doi: 10.1093/nar/gkt391. Epub 2013 May 15. Nucleic Acids Res. 2013. PMID: 23677608 Free PMC article.
-
BAGEL: a web-based bacteriocin genome mining tool.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W273-9. doi: 10.1093/nar/gkl237. Nucleic Acids Res. 2006. PMID: 16845009 Free PMC article.
-
BAGEL2: mining for bacteriocins in genomic data.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W647-51. doi: 10.1093/nar/gkq365. Epub 2010 May 12. Nucleic Acids Res. 2010. PMID: 20462861 Free PMC article.
-
In Silico Screening of Bacteriocin Gene Clusters within a Set of Marine Bacillota Genomes.Int J Mol Sci. 2024 Feb 22;25(5):2566. doi: 10.3390/ijms25052566. Int J Mol Sci. 2024. PMID: 38473813 Free PMC article.
-
[Recent advances in lanthipeptide biosynthesis - A review].Wei Sheng Wu Xue Bao. 2016 Mar 4;56(3):373-82. Wei Sheng Wu Xue Bao. 2016. PMID: 27382781 Review. Chinese.
Cited by
-
Draft Genome Sequence of Bacillus subtilis YBS29, a Potential Fish Probiotic That Prevents Motile Aeromonas Septicemia in Labeo rohita.Microbiol Resour Announc. 2022 Oct 20;11(10):e0091522. doi: 10.1128/mra.00915-22. Epub 2022 Sep 26. Microbiol Resour Announc. 2022. PMID: 36154193 Free PMC article.
-
Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs).Synth Syst Biotechnol. 2020 Jun 24;5(3):155-172. doi: 10.1016/j.synbio.2020.06.002. eCollection 2020 Sep. Synth Syst Biotechnol. 2020. PMID: 32637669 Free PMC article. Review.
-
Whole genome sequence analysis of Ligilactobacillus agilis C7 isolated from pig feces revealed three bacteriocin gene clusters.J Anim Sci Technol. 2022 Sep;64(5):1008-1011. doi: 10.5187/jast.2022.e55. Epub 2022 Sep 30. J Anim Sci Technol. 2022. PMID: 36287741 Free PMC article.
-
Production of Pumilarin and a Novel Circular Bacteriocin, Altitudin A, by Bacillus altitudinis ECC22, a Soil-Derived Bacteriocin Producer.Int J Mol Sci. 2024 Feb 7;25(4):2020. doi: 10.3390/ijms25042020. Int J Mol Sci. 2024. PMID: 38396696 Free PMC article.
-
Genome mining strategies for ribosomally synthesised and post-translationally modified peptides.Comput Struct Biotechnol J. 2020 Jun 25;18:1838-1851. doi: 10.1016/j.csbj.2020.06.032. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32728407 Free PMC article. Review.
References
-
- Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., Camarero J.A., Campopiano D.J., Challis G.L., Clardy J. et al. . Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013; 30:108–160. - PMC - PubMed
-
- Alt S., Wilkinson B.. Biosynthesis of the novel macrolide antibiotic anthracimycin. ACS Chem. Biol. 2015; 10:2468–2479. - PubMed
-
- van Heel A.J., Kloosterman T.G., Montalban-Lopez M., Deng J.-J., Plat A., Baudu B., Hendriks D., Moll G.N., Kuipers O.P.. Discovery, production and modification of 5 novel lantibiotics using the promiscuous nisin modification machinery. ACS Synth. Biol. 2016; 5:1146–1154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

