R-loops promote trinucleotide repeat deletion through DNA base excision repair enzymatic activities
- PMID: 32763971
- PMCID: PMC7535915
- DOI: 10.1074/jbc.RA120.014161
R-loops promote trinucleotide repeat deletion through DNA base excision repair enzymatic activities
Abstract
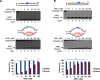
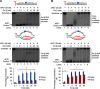
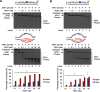
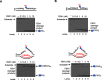
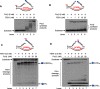
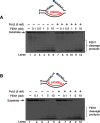
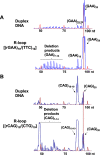
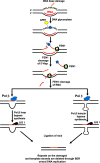
Trinucleotide repeat (TNR) expansion and deletion are responsible for over 40 neurodegenerative diseases and associated with cancer. TNRs can undergo somatic instability that is mediated by DNA damage and repair and gene transcription. Recent studies have pointed toward a role for R-loops in causing TNR expansion and deletion, and it has been shown that base excision repair (BER) can result in CAG repeat deletion from R-loops in yeast. However, it remains unknown how BER in R-loops can mediate TNR instability. In this study, using biochemical approaches, we examined BER enzymatic activities and their influence on TNR R-loops. We found that AP endonuclease 1 incised an abasic site on the nontemplate strand of a TNR R-loop, creating a double-flap intermediate containing an RNA:DNA hybrid that subsequently inhibited polymerase β (pol β) synthesis of TNRs. This stimulated flap endonuclease 1 (FEN1) cleavage of TNRs engaged in an R-loop. Moreover, we showed that FEN1 also efficiently cleaved the RNA strand, facilitating pol β loop/hairpin bypass synthesis and the resolution of TNR R-loops through BER. Consequently, this resulted in fewer TNRs synthesized by pol β than those removed by FEN1, thereby leading to repeat deletion. Our results indicate that TNR R-loops preferentially lead to repeat deletion during BER by disrupting the balance between the addition and removal of TNRs. Our discoveries open a new avenue for the treatment and prevention of repeat expansion diseases and cancer.
Keywords: DNA base damage; DNA damage; DNA endonuclease; DNA polymerase; DNA repair; DNA structure; R-loops; base excision repair; trinucleotide repeat instability.
© 2020 Laverde et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
Proliferating cell nuclear antigen prevents trinucleotide repeat expansions by promoting repeat deletion and hairpin removal.DNA Repair (Amst). 2016 Dec;48:17-29. doi: 10.1016/j.dnarep.2016.10.006. Epub 2016 Oct 22. DNA Repair (Amst). 2016. PMID: 27793507 Free PMC article.
-
Base excision repair of oxidative DNA damage coupled with removal of a CAG repeat hairpin attenuates trinucleotide repeat expansion.Nucleic Acids Res. 2014 Apr;42(6):3675-91. doi: 10.1093/nar/gkt1372. Epub 2014 Jan 14. Nucleic Acids Res. 2014. PMID: 24423876 Free PMC article.
-
Flap Endonuclease 1 Endonucleolytically Processes RNA to Resolve R-Loops through DNA Base Excision Repair.Genes (Basel). 2022 Dec 29;14(1):98. doi: 10.3390/genes14010098. Genes (Basel). 2022. PMID: 36672839 Free PMC article.
-
Trinucleotide repeat instability via DNA base excision repair.DNA Repair (Amst). 2020 Sep;93:102912. doi: 10.1016/j.dnarep.2020.102912. DNA Repair (Amst). 2020. PMID: 33087278 Free PMC article. Review.
-
DNA base excision repair: a mechanism of trinucleotide repeat expansion.Trends Biochem Sci. 2012 Apr;37(4):162-72. doi: 10.1016/j.tibs.2011.12.002. Epub 2012 Jan 27. Trends Biochem Sci. 2012. PMID: 22285516 Free PMC article. Review.
Cited by
-
Profiling human pathogenic repeat expansion regions by synergistic and multi-level impacts on molecular connections.Hum Genet. 2023 Feb;142(2):245-274. doi: 10.1007/s00439-022-02500-6. Epub 2022 Nov 7. Hum Genet. 2023. PMID: 36344696 Free PMC article.
-
Role of Senataxin in Amyotrophic Lateral Sclerosis.J Mol Neurosci. 2023 Dec;73(11-12):996-1009. doi: 10.1007/s12031-023-02169-0. Epub 2023 Nov 20. J Mol Neurosci. 2023. PMID: 37982993 Review.
-
Walking a tightrope: The complex balancing act of R-loops in genome stability.Mol Cell. 2022 Jun 16;82(12):2267-2297. doi: 10.1016/j.molcel.2022.04.014. Epub 2022 May 3. Mol Cell. 2022. PMID: 35508167 Free PMC article. Review.
-
PCNA Ser46-Leu47 residues are crucial in preserving genomic integrity.PLoS One. 2023 May 19;18(5):e0285337. doi: 10.1371/journal.pone.0285337. eCollection 2023. PLoS One. 2023. PMID: 37205694 Free PMC article.
-
Does the XPA-FEN1 Interaction Concern to Nucleotide Excision Repair or Beyond?Biomolecules. 2024 Jul 9;14(7):814. doi: 10.3390/biom14070814. Biomolecules. 2024. PMID: 39062528 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

