The Branched Nature of the Nonsense-Mediated mRNA Decay Pathway
- PMID: 33008628
- PMCID: PMC7854845
- DOI: 10.1016/j.tig.2020.08.010
The Branched Nature of the Nonsense-Mediated mRNA Decay Pathway
Abstract
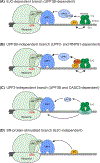
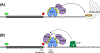
Nonsense-mediated mRNA decay (NMD) is a conserved translation-coupled quality control mechanism in all eukaryotes that regulates the expression of a significant fraction of both the aberrant and normal transcriptomes. In vertebrates, NMD has become an essential process owing to expansion of the diversity of NMD-regulated transcripts, particularly during various developmental processes. Surprisingly, however, some core NMD factors that are essential for NMD in simpler organisms appear to be dispensable for vertebrate NMD. At the same time, numerous NMD enhancers and suppressors have been identified in multicellular organisms including vertebrates. Collectively, the available data suggest that vertebrate NMD is a complex, branched pathway wherein individual branches regulate specific mRNA subsets to fulfill distinct physiological functions.
Keywords: 3′-UTR; UPF proteins; exon-junction complex; mRNA decay; nonsense codons; premature termination codons.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
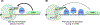
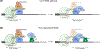

Similar articles
-
Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes.Nat Rev Mol Cell Biol. 2015 Nov;16(11):665-77. doi: 10.1038/nrm4063. Epub 2015 Sep 23. Nat Rev Mol Cell Biol. 2015. PMID: 26397022 Review.
-
Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways.RNA. 2013 Oct;19(10):1432-48. doi: 10.1261/rna.038893.113. Epub 2013 Aug 20. RNA. 2013. PMID: 23962664 Free PMC article.
-
The Meaning of NMD: Translate or Perish.Trends Genet. 2016 Jul;32(7):395-407. doi: 10.1016/j.tig.2016.04.007. Epub 2016 May 14. Trends Genet. 2016. PMID: 27185236 Review.
-
Arginine CGA codons as a source of nonsense mutations: a possible role in multivariant gene expression, control of mRNA quality, and aging.Mol Genet Genomics. 2017 Oct;292(5):1013-1026. doi: 10.1007/s00438-017-1328-y. Epub 2017 May 18. Mol Genet Genomics. 2017. PMID: 28523359
-
Mechanism and regulation of the nonsense-mediated decay pathway.Nucleic Acids Res. 2016 Feb 29;44(4):1483-95. doi: 10.1093/nar/gkw010. Epub 2016 Jan 14. Nucleic Acids Res. 2016. PMID: 26773057 Free PMC article. Review.
Cited by
-
Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection.Mol Cell. 2021 Jul 1;81(13):2851-2867.e7. doi: 10.1016/j.molcel.2021.05.023. Epub 2021 May 24. Mol Cell. 2021. PMID: 34118193 Free PMC article.
-
An alternative UPF1 isoform drives conditional remodeling of nonsense-mediated mRNA decay.EMBO J. 2022 May 16;41(10):e108898. doi: 10.15252/embj.2021108898. Epub 2022 Apr 11. EMBO J. 2022. PMID: 35403729 Free PMC article.
-
Regulation of Early Lymphocyte Development via mRNA Decay Catalyzed by the CCR4-NOT Complex.Front Immunol. 2021 Jul 19;12:715675. doi: 10.3389/fimmu.2021.715675. eCollection 2021. Front Immunol. 2021. PMID: 34349771 Free PMC article. Review.
-
Spatial expression of the nonsense-mediated mRNA decay factors UPF3A and UPF3B among mouse tissues.J Zhejiang Univ Sci B. 2023 Nov 15;24(11):1062-1068. doi: 10.1631/jzus.B2300126. J Zhejiang Univ Sci B. 2023. PMID: 37961809 Free PMC article.
-
Partial sequence identity in a 25-nucleotide long element is sufficient for transcriptional adaptation in the Caenorhabditis elegans act-5/act-3 model.PLoS Genet. 2023 Jun 29;19(6):e1010806. doi: 10.1371/journal.pgen.1010806. eCollection 2023 Jun. PLoS Genet. 2023. PMID: 37384903 Free PMC article.
References
-
- Mendell JT et al. (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet 36, 1073–1078 - PubMed
-
- Pulak R and Anderson P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7, 1885–1897 - PubMed
-
- Medghalchi SM et al. (2001) Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet 10, 99–105 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

