Inhibition of the SUV4-20 H1 histone methyltransferase increases frataxin expression in Friedreich's ataxia patient cells
- PMID: 33028632
- PMCID: PMC7939392
- DOI: 10.1074/jbc.RA120.015533
Inhibition of the SUV4-20 H1 histone methyltransferase increases frataxin expression in Friedreich's ataxia patient cells
Abstract
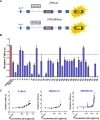
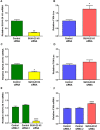
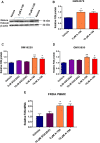
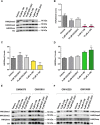
The molecular mechanisms of reduced frataxin (FXN) expression in Friedreich's ataxia (FRDA) are linked to epigenetic modification of the FXN locus caused by the disease-associated GAA expansion. Here, we identify that SUV4-20 histone methyltransferases, specifically SUV4-20 H1, play an important role in the regulation of FXN expression and represent a novel therapeutic _target. Using a human FXN-GAA-Luciferase repeat expansion genomic DNA reporter model of FRDA, we screened the Structural Genomics Consortium epigenetic probe collection. We found that pharmacological inhibition of the SUV4-20 methyltransferases by the tool compound A-196 increased the expression of FXN by ∼1.5-fold in the reporter cell line. In several FRDA cell lines and patient-derived primary peripheral blood mononuclear cells, A-196 increased FXN expression by up to 2-fold, an effect not seen in WT cells. SUV4-20 inhibition was accompanied by a reduction in H4K20me2 and H4K20me3 and an increase in H4K20me1, but only modest (1.4-7.8%) perturbation in genome-wide expression was observed. Finally, based on the structural activity relationship and crystal structure of A-196, novel small molecule A-196 analogs were synthesized and shown to give a 20-fold increase in potency for increasing FXN expression. Overall, our results suggest that histone methylation is important in the regulation of FXN expression and highlight SUV4-20 H1 as a potential novel therapeutic _target for FRDA.
Keywords: Friedreich's ataxia; drug screening; epigenetics; frataxin; histone methylation.
© 2020 Vilema-Enríquez et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures
Similar articles
-
A GAA repeat expansion reporter model of Friedreich's ataxia recapitulates the genomic context and allows rapid screening of therapeutic compounds.Hum Mol Genet. 2013 Dec 20;22(25):5173-87. doi: 10.1093/hmg/ddt370. Epub 2013 Aug 13. Hum Mol Genet. 2013. PMID: 23943791 Free PMC article.
-
Mitochondrial damage and senescence phenotype of cells derived from a novel frataxin G127V point mutation mouse model of Friedreich's ataxia.Dis Model Mech. 2020 Jul 27;13(7):dmm045229. doi: 10.1242/dmm.045229. Dis Model Mech. 2020. PMID: 32586831 Free PMC article.
-
Alleviating GAA Repeat Induced Transcriptional Silencing of the Friedreich's Ataxia Gene During Somatic Cell Reprogramming.Stem Cells Dev. 2016 Dec 1;25(23):1788-1800. doi: 10.1089/scd.2016.0147. Epub 2016 Oct 17. Stem Cells Dev. 2016. PMID: 27615158 Free PMC article.
-
Increasing frataxin gene expression with histone deacetylase inhibitors as a therapeutic approach for Friedreich's ataxia.J Neurochem. 2013 Aug;126 Suppl 1(0 1):147-54. doi: 10.1111/jnc.12302. J Neurochem. 2013. PMID: 23859350 Free PMC article. Review.
-
Gene regulation and epigenetics in Friedreich's ataxia.J Neurochem. 2013 Aug;126 Suppl 1:21-42. doi: 10.1111/jnc.12254. J Neurochem. 2013. PMID: 23859339 Review.
Cited by
-
The SUV4-20H Histone Methyltransferases in Health and Disease.Int J Mol Sci. 2022 Apr 25;23(9):4736. doi: 10.3390/ijms23094736. Int J Mol Sci. 2022. PMID: 35563127 Free PMC article. Review.
-
Recessive cerebellar and afferent ataxias - clinical challenges and future directions.Nat Rev Neurol. 2022 May;18(5):257-272. doi: 10.1038/s41582-022-00634-9. Epub 2022 Mar 24. Nat Rev Neurol. 2022. PMID: 35332317 Review.
-
DNA Base Damage Repair Crosstalks with Chromatin Structures to Contract Expanded GAA Repeats in Friedreich's Ataxia.Biomolecules. 2024 Jul 8;14(7):809. doi: 10.3390/biom14070809. Biomolecules. 2024. PMID: 39062522 Free PMC article.
-
The fragile X locus is prone to spontaneous DNA damage that is preferentially repaired by nonhomologous end-joining to preserve genome integrity.iScience. 2024 Jan 6;27(2):108814. doi: 10.1016/j.isci.2024.108814. eCollection 2024 Feb 16. iScience. 2024. PMID: 38303711 Free PMC article.
-
Selected Histone Deacetylase Inhibitors Reverse the Frataxin Transcriptional Defect in a Novel Friedreich's Ataxia Induced Pluripotent Stem Cell-Derived Neuronal Reporter System.Front Neurosci. 2022 Feb 23;16:836476. doi: 10.3389/fnins.2022.836476. eCollection 2022. Front Neurosci. 2022. PMID: 35281493 Free PMC article.
References
-
- Campuzano V., Montermini L., Moltò M.D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Sanjay I., Gellera C. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. 8596916. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

