sRNAbench and sRNAtoolbox 2022 update: accurate miRNA and sncRNA profiling for model and non-model organisms
- PMID: 35556129
- PMCID: PMC9252802
- DOI: 10.1093/nar/gkac363
sRNAbench and sRNAtoolbox 2022 update: accurate miRNA and sncRNA profiling for model and non-model organisms
Abstract
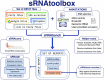
The NCBI Sequence Read Archive currently hosts microRNA sequencing data for over 800 different species, evidencing the existence of a broad taxonomic distribution in the field of small RNA research. Simultaneously, the number of samples per miRNA-seq study continues to increase resulting in a vast amount of data that requires accurate, fast and user-friendly analysis methods. Since the previous release of sRNAtoolbox in 2019, 55 000 sRNAbench jobs have been submitted which has motivated many improvements in its usability and the scope of the underlying annotation database. With this update, users can upload an unlimited number of samples or import them from Google Drive, Dropbox or URLs. Micro- and small RNA profiling can now be carried out using high-confidence Metazoan and plant specific databases, MirGeneDB and PmiREN respectively, together with genome assemblies and libraries from 441 Ensembl species. The new results page includes straightforward sample annotation to allow downstream differential expression analysis with sRNAde. Unassigned reads can also be explored by means of a new tool that performs mapping to microbial references, which can reveal contamination events or biologically meaningful findings as we describe in the example. sRNAtoolbox is available at: https://arn.ugr.es/srnatoolbox/.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
sRNAbench and sRNAtoolbox 2019: intuitive fast small RNA profiling and differential expression.Nucleic Acids Res. 2019 Jul 2;47(W1):W530-W535. doi: 10.1093/nar/gkz415. Nucleic Acids Res. 2019. PMID: 31114926 Free PMC article.
-
sRNAtoolbox: Dockerized Analysis of Small RNA Sequencing Data in Model and Non-model Species.Methods Mol Biol. 2023;2630:179-213. doi: 10.1007/978-1-0716-2982-6_13. Methods Mol Biol. 2023. PMID: 36689184
-
sRNAtoolbox: an integrated collection of small RNA research tools.Nucleic Acids Res. 2015 Jul 1;43(W1):W467-73. doi: 10.1093/nar/gkv555. Epub 2015 May 27. Nucleic Acids Res. 2015. PMID: 26019179 Free PMC article.
-
Computational tools supporting known miRNA identification.Prog Mol Biol Transl Sci. 2024;203:225-242. doi: 10.1016/bs.pmbts.2023.12.018. Epub 2024 Jan 30. Prog Mol Biol Transl Sci. 2024. PMID: 38360000 Review.
-
A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome.Annu Rev Genet. 2015;49:213-42. doi: 10.1146/annurev-genet-120213-092023. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473382 Free PMC article. Review.
Cited by
-
RNA-seq validation of microRNA expression signatures for precision melanoma diagnosis and prognostic stratification.BMC Med Genomics. 2024 Oct 25;17(1):256. doi: 10.1186/s12920-024-02028-w. BMC Med Genomics. 2024. PMID: 39456086 Free PMC article.
-
A glimpse into the world of microRNAs and their putative roles in hard ticks.Front Cell Dev Biol. 2024 Sep 23;12:1460705. doi: 10.3389/fcell.2024.1460705. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39376631 Free PMC article. Review.
-
Delimitation of the widely distributed Palearctic Stenodema species (Hemiptera, Heteroptera, Miridae): insights from molecular and morphological data.Zookeys. 2024 Aug 13;1209:245-294. doi: 10.3897/zookeys.1209.124766. eCollection 2024. Zookeys. 2024. PMID: 39175835 Free PMC article.
-
In search of epigenetic hallmarks of different tissues: an integrative omics study of horse liver, lung, and heart.Mamm Genome. 2024 Dec;35(4):600-620. doi: 10.1007/s00335-024-10057-0. Epub 2024 Aug 14. Mamm Genome. 2024. PMID: 39143382 Free PMC article.
-
Analysis of Amblyomma americanum microRNAs in response to Ehrlichia chaffeensis infection and their potential role in vectorial capacity.Front Cell Infect Microbiol. 2024 Jul 17;14:1427562. doi: 10.3389/fcimb.2024.1427562. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39086604 Free PMC article.
References
-
- Lu C., Meyers B.C., Green P.J.. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007; 43:110–117. - PubMed

