miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach
- PMID: 36499183
- PMCID: PMC9739077
- DOI: 10.3390/ijms232314855
miR-18a-5p Is Involved in the Developmental Origin of Prostate Cancer in Maternally Malnourished Offspring Rats: A DOHaD Approach
Abstract
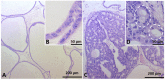
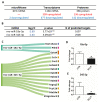
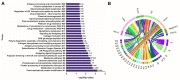
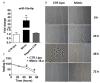
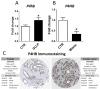
The Developmental Origins of Health and Disease (DOHaD) concept correlates early life exposure to stressor conditions with the increased incidence of non-communicable chronic diseases, including prostate cancer (PCa), throughout the life span. However, the molecular mechanisms involved in this process remain poorly understood. In this study, the deregulation of two miRNAs (rno-miR-18a-5p and rno-miR-345-3p) was described in the ventral prostate VP of old rats born to dams fed with a low protein diet (LPD) (6% protein in the diet) during gestational and lactational periods. Integrative analysis of the (VP) transcriptomic and proteomic data revealed changes in the expression profile of 14 identified predicted _targets of these two DE miRNAs, which enriched terms related to post-translational protein modification, metabolism of proteins, protein processing in endoplasmic reticulum, phosphonate and phosphinate metabolism, the calnexin/calreticulin cycle, metabolic pathways, N-glycan trimming in the ER and the calnexin/calreticulin cycle, hedgehog ligand biogenesis, the ER-phagosome pathway, detoxification of reactive oxygen species, antigenprocessing-cross presentation, RAB geranylgeranylation, collagen formation, glutathione metabolism, the metabolism of xenobiotics by cytochrome P450, and platinum drug resistance. RT-qPCR validated the deregulation of the miR-18a-5p/P4HB (prolyl 4-hydroxylase subunit beta) network in the VP of older offspring as well as in the PNT-2 cells transfected with mimic miR-18a-5p. Functional in vitro studies revealed a potential modulation of estrogen receptor α (ESR1) by miR-18a-5p in PNT-2 cells, which was also confirmed in the VP of older offspring. An imbalance of the testosterone/estrogen ratio was also observed in the offspring rats born to dams fed with an LPD. In conclusion, deregulation of the miR-18a-5p/P4HB network can contribute to the developmental origins of prostate cancer in maternally malnourished offspring, highlighting the need for improving maternal healthcare during critical windows of vulnerability early in life.
Keywords: Developmental Origins of Health and Disease (DOHaD); aging; epigenetics; miR-18a-5p/P4HB network; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
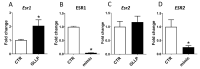
Similar articles
-
Early-life origin of prostate cancer through deregulation of miR-206 networks in maternally malnourished offspring rats.Sci Rep. 2023 Oct 31;13(1):18685. doi: 10.1038/s41598-023-46068-1. Sci Rep. 2023. PMID: 37907720 Free PMC article.
-
Deregulation of ABCG1 early in life contributes to prostate carcinogenesis in maternally malnourished offspring rats.Mol Cell Endocrinol. 2024 Jan 15;580:112102. doi: 10.1016/j.mce.2023.112102. Epub 2023 Nov 14. Mol Cell Endocrinol. 2024. PMID: 37972683
-
In silico investigation of the role of miRNAs in a possible developmental origin of prostate cancer in F1 and F2 offspring of mothers exposed to a phthalate mixture.Environ Toxicol. 2024 Jun;39(6):3523-3536. doi: 10.1002/tox.24181. Epub 2024 Mar 11. Environ Toxicol. 2024. PMID: 38465474
-
Deep RNA sequencing reveals that microRNAs play a key role in lactation in rats.J Nutr. 2014 Aug;144(8):1142-9. doi: 10.3945/jn.114.192575. Epub 2014 Jun 4. J Nutr. 2014. PMID: 24899157
-
Activin A regulates microRNAs and gene expression in LNCaP cells.Prostate. 2016 Aug;76(11):951-63. doi: 10.1002/pros.23184. Epub 2016 Mar 28. Prostate. 2016. PMID: 27018851
Cited by
-
Early-life origin of prostate cancer through deregulation of miR-206 networks in maternally malnourished offspring rats.Sci Rep. 2023 Oct 31;13(1):18685. doi: 10.1038/s41598-023-46068-1. Sci Rep. 2023. PMID: 37907720 Free PMC article.
References
-
- Newman C.B., Preiss D., Tobert J.A., Jacobson T.A., Page I.R.L., Goldstein L.B., Chin C., Tannock L.R., Miller M., Raghuveer G., et al. Statin Safety and Associated Adverse Events: A Scientific Statement From the American Heart Association. Arter. Thromb. Vasc. Biol. 2019;39:e38–e81. doi: 10.1161/ATV.0000000000000073. - DOI - PubMed
-
- McKerracher L., Moffat T., Barker M., McConnell M., Atkinson S.A., Murray-Davis B., McDonald S.D., Sloboda D.M. Knowledge about the Developmental Origins of Health and Disease is independently associated with variation in diet quality during pregnancy. Matern. Child Nutr. 2019;16:e12891. doi: 10.1111/mcn.12891. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

